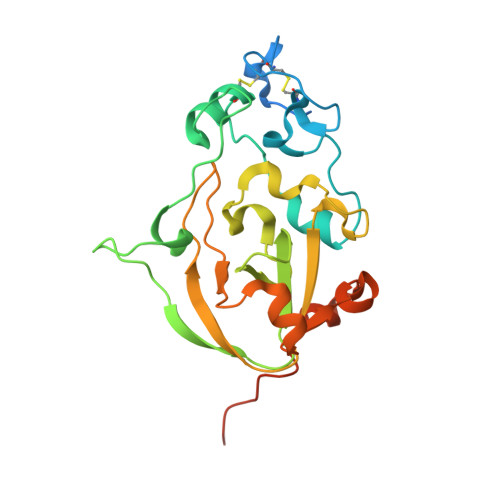
Discovery of fungal surface NADases predominantly present in pathogenic species.
Stromland, O., Kallio, J.P., Pschibul, A., Skoge, R.H., Hardardottir, H.M., Sverkeli, L.J., Heinekamp, T., Kniemeyer, O., Migaud, M., Makarov, M.V., Gossmann, T.I., Brakhage, A.A., Ziegler, M.(2021) Nat Commun 12: 1631-1631
- PubMed: 33712585
- DOI: https://doi.org/10.1038/s41467-021-21307-z
- Primary Citation of Related Structures:
6YGE, 6YGF, 6YGG - PubMed Abstract:
Nicotinamide adenine dinucleotide (NAD) is a key molecule in cellular bioenergetics and signalling. Various bacterial pathogens release NADase enzymes into the host cell that deplete the host's NAD + pool, thereby causing rapid cell death. Here, we report the identification of NADases on the surface of fungi such as the pathogen Aspergillus fumigatus and the saprophyte Neurospora crassa. The enzymes harbour a tuberculosis necrotizing toxin (TNT) domain and are predominately present in pathogenic species. The 1.6 Å X-ray structure of the homodimeric A. fumigatus protein reveals unique properties including N-linked glycosylation and a Ca 2+ -binding site whose occupancy regulates activity. The structure in complex with a substrate analogue suggests a catalytic mechanism that is distinct from those of known NADases, ADP-ribosyl cyclases and transferases. We propose that fungal NADases may convey advantages during interaction with the host or competing microorganisms.
- Department of Biomedicine, University of Bergen, Bergen, Norway.
Organizational Affiliation: