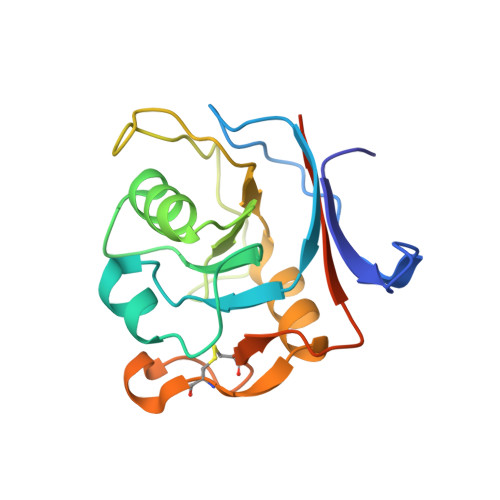
Computational backbone design enables soluble engineering of transferrin receptor apical domain.
Sjostrom, D.J., Berger, S.A., Oberdorfer, G., Bjelic, S.(2020) Proteins 88: 1569-1577
- PubMed: 32592192
- DOI: https://doi.org/10.1002/prot.25974
- Primary Citation of Related Structures:
6Y76 - PubMed Abstract:
Supply of iron into human cells is achieved by iron carrier protein transferrin and its receptor that upon complex formation get internalized by endocytosis. Similarly, the iron needs to be delivered into the brain, and necessitates the transport across the blood-brain barrier. While there are still unanswered questions about these mechanisms, extensive efforts have been made to use the system for delivery of therapeutics into biological compartments. The dimeric form of the receptor, where each subunit consists of three domains, further complicates the detailed investigation of molecular determinants responsible for guiding the receptor interactions with other proteins. Especially the apical domain's biological function has been elusive. To further the study of transferrin receptor, we have computationally decoupled the apical domain for soluble expression, and validated the design strategy by structure determination. Besides presenting a methodology for solubilizing domains, the results will allow for study of apical domain's function.
- Department of Chemistry and Biomedical Sciences, Linnaeus University, Kalmar, Sweden.
Organizational Affiliation: