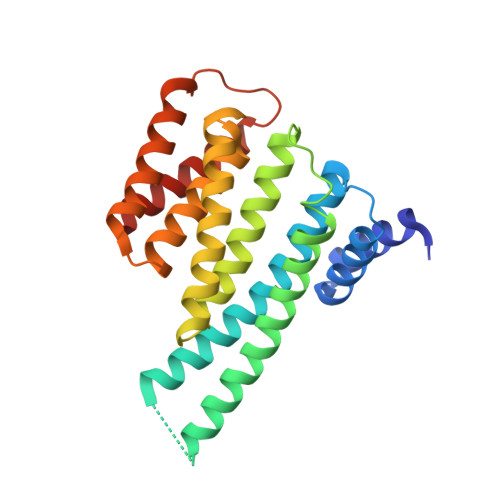
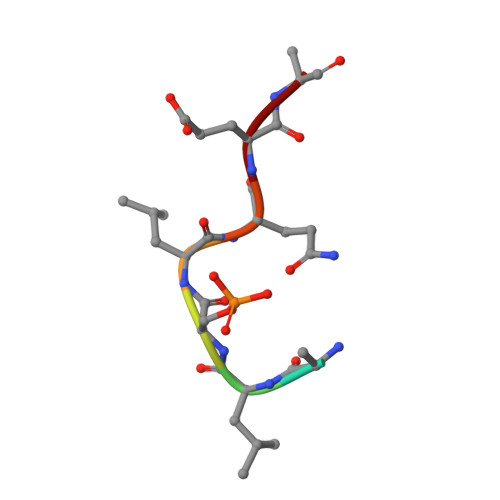
Stabilization of Protein-Protein Interactions between CaMKK2 and 14-3-3 by Fusicoccins.
Lentini Santo, D., Petrvalska, O., Obsilova, V., Ottmann, C., Obsil, T.(2020) ACS Chem Biol 15: 3060-3071
- PubMed: 33146997
- DOI: https://doi.org/10.1021/acschembio.0c00821
- Primary Citation of Related Structures:
6Y4K, 6Y6B - PubMed Abstract:
Ca 2+ /calmodulin-dependent protein kinase kinase 2 (CaMKK2) regulates several key physiological and pathophysiological processes, and its dysregulation has been implicated in obesity, diabetes, and cancer. CaMKK2 is inhibited through phosphorylation in a process involving binding to the scaffolding 14-3-3 protein, which maintains CaMKK2 in the phosphorylation-mediated inhibited state. The previously reported structure of the N-terminal CaMKK2 14-3-3-binding motif bound to 14-3-3 suggested that the interaction between 14-3-3 and CaMKK2 could be stabilized by small-molecule compounds. Thus, we investigated the stabilization of interactions between CaMKK2 and 14-3-3γ by Fusicoccin A and other fusicoccanes-diterpene glycosides that bind at the interface between the 14-3-3 ligand binding groove and the 14-3-3 binding motif of the client protein. Our data reveal that two of five tested fusicoccanes considerably increase the binding of phosphopeptide representing the 14-3-3 binding motif of CaMKK2 to 14-3-3γ. Crystal structures of two ternary complexes suggest that the steric contacts between the C-terminal part of the CaMKK2 14-3-3 binding motif and the adjacent fusicoccane molecule are responsible for differences in stabilization potency between the study compounds. Moreover, our data also show that fusicoccanes enhance the binding affinity of phosphorylated full-length CaMKK2 to 14-3-3γ, which in turn slows down CaMKK2 dephosphorylation, thus keeping this protein in its phosphorylation-mediated inhibited state. Therefore, targeting the fusicoccin binding cavity of 14-3-3 by small-molecule compounds may offer an alternative strategy to suppress CaMKK2 activity by stabilizing its phosphorylation-mediated inhibited state.
- Department of Physical and Macromolecular Chemistry, Faculty of Science, Charles University, Prague, Czech Republic.
Organizational Affiliation: