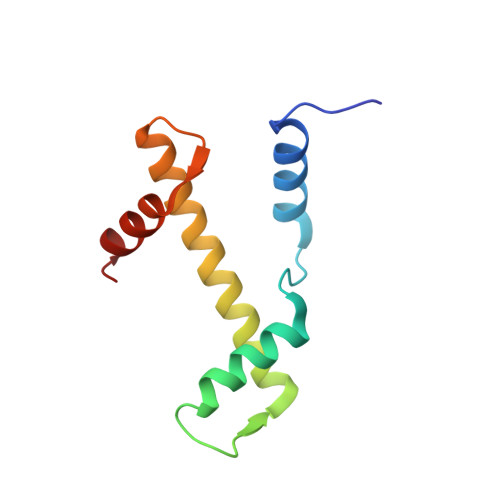
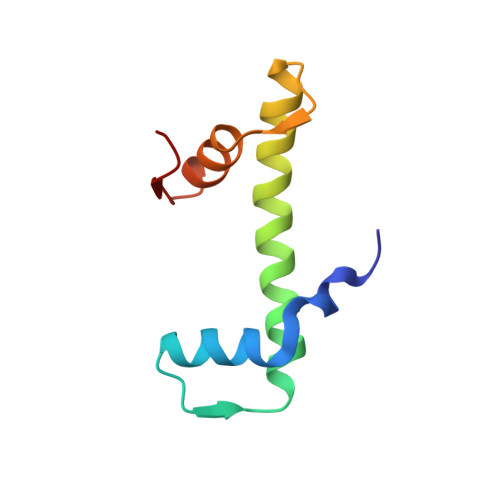
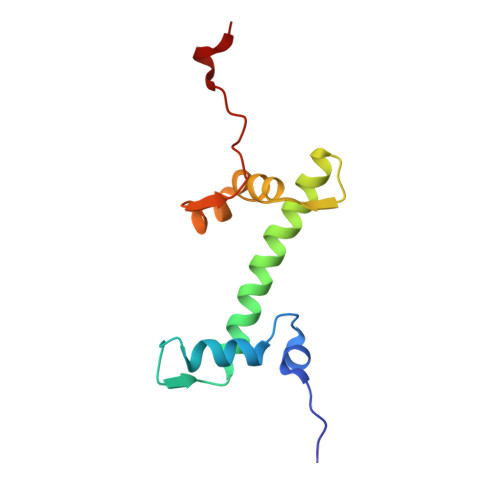
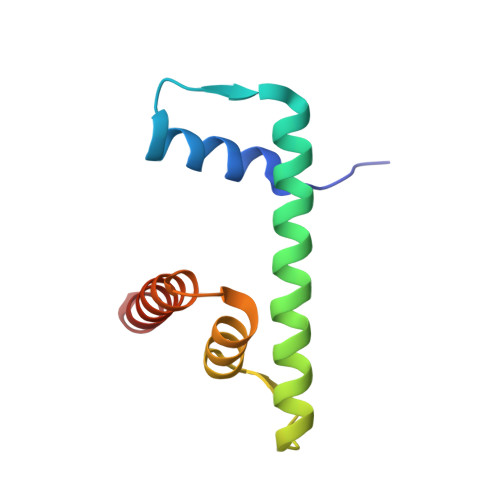
Structural mechanism of cGAS inhibition by the nucleosome.
Pathare, G.R., Decout, A., Gluck, S., Cavadini, S., Makasheva, K., Hovius, R., Kempf, G., Weiss, J., Kozicka, Z., Guey, B., Melenec, P., Fierz, B., Thoma, N.H., Ablasser, A.(2020) Nature 587: 668-672
- PubMed: 32911482
- DOI: https://doi.org/10.1038/s41586-020-2750-6
- Primary Citation of Related Structures:
6Y5D, 6Y5E - PubMed Abstract:
The DNA sensor cyclic GMP-AMP synthase (cGAS) initiates innate immune responses following microbial infection, cellular stress and cancer 1 . Upon activation by double-stranded DNA, cytosolic cGAS produces 2'3' cGMP-AMP, which triggers the induction of inflammatory cytokines and type I interferons 2-7 . cGAS is also present inside the cell nucleus, which is replete with genomic DNA 8 , where chromatin has been implicated in restricting its enzymatic activity 9 . However, the structural basis for inhibition of cGAS by chromatin remains unknown. Here we present the cryo-electron microscopy structure of human cGAS bound to nucleosomes. cGAS makes extensive contacts with both the acidic patch of the histone H2A-H2B heterodimer and nucleosomal DNA. The structural and complementary biochemical analysis also find cGAS engaged to a second nucleosome in trans. Mechanistically, binding of the nucleosome locks cGAS into a monomeric state, in which steric hindrance suppresses spurious activation by genomic DNA. We find that mutations to the cGAS-acidic patch interface are sufficient to abolish the inhibitory effect of nucleosomes in vitro and to unleash the activity of cGAS on genomic DNA in living cells. Our work uncovers the structural basis of the interaction between cGAS and chromatin and details a mechanism that permits self-non-self discrimination of genomic DNA by cGAS.
- Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland.
Organizational Affiliation: