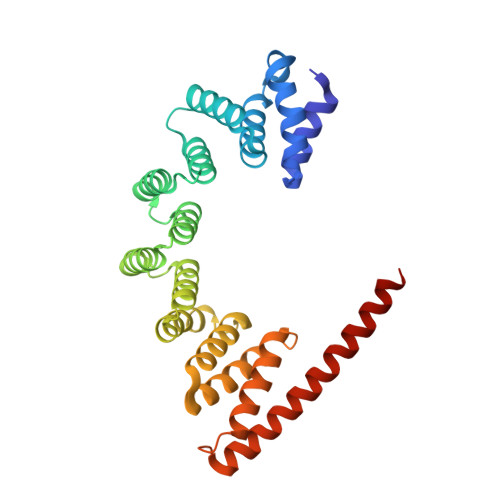
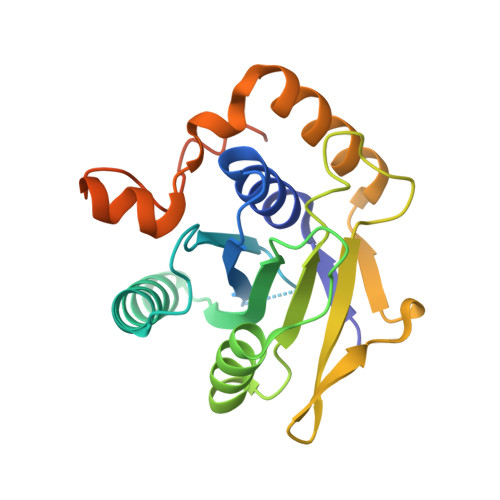
The architecture of EMC reveals a path for membrane protein insertion.
O'Donnell, J.P., Phillips, B.P., Yagita, Y., Juszkiewicz, S., Wagner, A., Malinverni, D., Keenan, R.J., Miller, E.A., Hegde, R.S.(2020) Elife 9
- PubMed: 32459176
- DOI: https://doi.org/10.7554/eLife.57887
- Primary Citation of Related Structures:
6Y4L, 6Z3W - PubMed Abstract:
Approximately 25% of eukaryotic genes code for integral membrane proteins that are assembled at the endoplasmic reticulum. An abundant and widely conserved multi-protein complex termed EMC has been implicated in membrane protein biogenesis, but its mechanism of action is poorly understood. Here, we define the composition and architecture of human EMC using biochemical assays, crystallography of individual subunits, site-specific photocrosslinking, and cryo-EM reconstruction. Our results suggest that EMC's cytosolic domain contains a large, moderately hydrophobic vestibule that can bind a substrate's transmembrane domain (TMD). The cytosolic vestibule leads into a lumenally-sealed, lipid-exposed intramembrane groove large enough to accommodate a single substrate TMD. A gap between the cytosolic vestibule and intramembrane groove provides a potential path for substrate egress from EMC. These findings suggest how EMC facilitates energy-independent membrane insertion of TMDs, explain why only short lumenal domains are translocated by EMC, and constrain models of EMC's proposed chaperone function.
- MRC Laboratory of Molecular Biology, Cambridge, United Kingdom.
Organizational Affiliation: