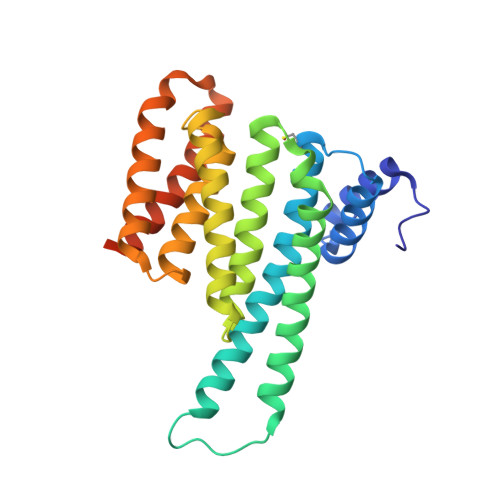
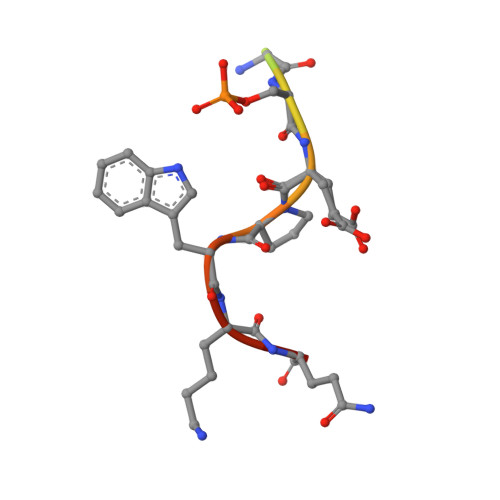
Interaction of an I kappa B alpha Peptide with 14-3-3.
Wolter, M., Santo, D.L., Herman, P., Ballone, A., Centorrino, F., Obsil, T., Ottmann, C.(2020) ACS Omega 5: 5380-5388
- PubMed: 32201828
- DOI: https://doi.org/10.1021/acsomega.9b04413
- Primary Citation of Related Structures:
6Y1J - PubMed Abstract:
Inflammatory responses mediated by the transcription factor nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB) play key roles in immunity, autoimmune diseases, and cancer. NF-κB is directly regulated through protein-protein interactions, including those with IκB and 14-3-3 proteins. These two important regulatory proteins have been reported to interact with each other, although little is known about this interaction. We analyzed the inhibitor of nuclear factor kappa B α (IκBα)/14-3-3σ interaction via a peptide/protein-based approach. Structural data were acquired via X-ray crystallography, while binding affinities were measured with fluorescence polarization assays and time-resolved tryptophan fluorescence. A high-resolution crystal structure (1.13 Å) of the uncommon 14-3-3 interaction motif of IκBα (IκBαpS63) in a complex with 14-3-3σ was evaluated. This motif harbors a tryptophan that makes this crystal structure the first one with such a residue visible in the electron density at that position. We used this tryptophan to determine the binding affinity of the unlabeled IκBα peptide to 14-3-3 via tryptophan fluorescence decay measurements.
- Department of Biomedical Engineering, Laboratory of Chemical Biology and Institute for Complex Molecular Systems, Eindhoven University of Technology, P.O. Box 513, 5600 MB Eindhoven, The Netherlands.
Organizational Affiliation: