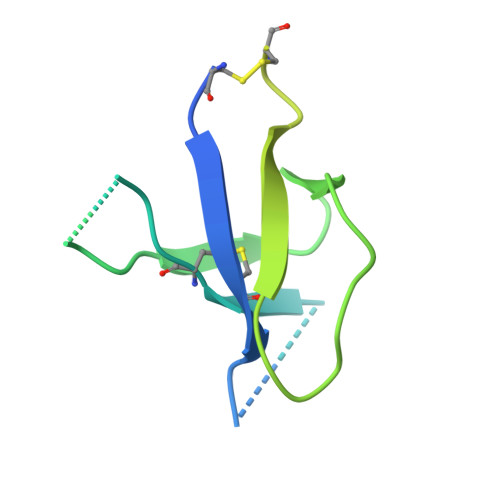
Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Aljedani, S.S., Liban, T.J., Tran, K., Phad, G., Singh, S., Dubrovskaya, V., Pushparaj, P., Martinez-Murillo, P., Rodarte, J., Mileant, A., Mangala Prasad, V., Kinzelman, R., O'Dell, S., Mascola, J.R., Lee, K.K., Karlsson Hedestam, G.B., Wyatt, R.T., Pancera, M.(2021) PLoS Pathog 17: e1009543-e1009543
- PubMed: 34559844
- DOI: https://doi.org/10.1371/journal.ppat.1009543
- Primary Citation of Related Structures:
6VJN, 6WAS, 6WIT, 6XLZ, 6XSN - PubMed Abstract:
Understanding the molecular mechanisms by which antibodies target and neutralize the HIV-1 envelope glycoprotein (Env) is critical in guiding immunogen design and vaccine development aimed at eliciting cross-reactive neutralizing antibodies (NAbs). Here, we analyzed monoclonal antibodies (mAbs) isolated from non-human primates (NHPs) immunized with variants of a native flexibly linked (NFL) HIV-1 Env stabilized trimer derived from the tier 2 clade C 16055 strain. The antibodies displayed neutralizing activity against the autologous virus with potencies ranging from 0.005 to 3.68 μg/ml (IC50). Structural characterization using negative-stain EM and X-ray crystallography identified the variable region 2 (V2) of the 16055 NFL trimer to be the common epitope for these antibodies. The crystal structures revealed that the V2 segment adopts a β-hairpin motif identical to that observed in the 16055 NFL crystal structure. These results depict how vaccine-induced antibodies derived from different clonal lineages penetrate through the glycan shield to recognize a hypervariable region within V2 (residues 184-186) that is unique to the 16055 strain. They also provide potential explanations for the potent autologous neutralization of these antibodies, confirming the immunodominance of this site and revealing that multiple angles of approach are permissible for affinity/avidity that results in potent neutralizing capacity. The structural analysis reveals that the most negatively charged paratope correlated with the potency of the mAbs. The atomic level information is of interest to both define the means of autologous neutralization elicited by different tier 2-based immunogens and facilitate trimer redesign to better target more conserved regions of V2 to potentially elicit cross-neutralizing HIV-1 antibodies.
- Fred Hutchinson Cancer Research Center, Vaccine and Infectious Disease Division, Seattle, Washington, United States of America.
Organizational Affiliation: