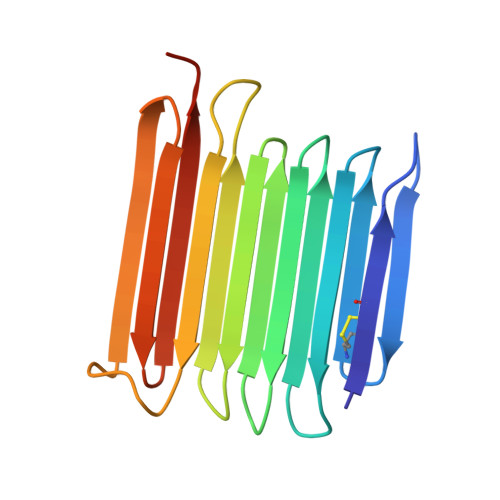
Crystal structure of an insect antifreeze protein reveals ordered waters on the ice-binding surface.
Ye, Q., Eves, R., Campbell, R.L., Davies, P.L.(2020) Biochem J 477: 3271-3286
- PubMed: 32794579
- DOI: https://doi.org/10.1042/BCJ20200539
- Primary Citation of Related Structures:
6XNR - PubMed Abstract:
Antifreeze proteins (AFPs) are characterized by their ability to adsorb to the surface of ice crystals and prevent any further crystal growth. AFPs have independently evolved for this purpose in a variety of organisms that encounter the threat of freezing, including many species of polar fish, insects, plants and microorganisms. Despite their diverse origins and structures, it has been suggested that all AFPs can organize ice-like water patterns on one side of the protein (the ice-binding site) that helps bind the AFP to ice. Here, to test this hypothesis, we have solved the crystal structure at 2.05 Å resolution of an AFP from the longhorn beetle, Rhagium mordax with five molecules in the unit cell. This AFP is hyperactive, and its crystal structure resembles that of the R. inquisitor ortholog in having a β-solenoid fold with a wide, flat ice-binding surface formed by four parallel rows of mainly Thr residues. The key difference between these structures is that the R. inquisitor AFP crystallized with its ice-binding site (IBS) making protein-protein contacts that limited the surface water patterns. Whereas the R. mordax AFP crystallized with the IBSs exposed to solvent enabling two layers of unrestricted ordered surface waters to be seen. These crystal waters make close matches to ice lattice waters on the basal and primary prism planes.
- Department of Biomedical and Molecular Sciences, Queen's University, Kingston, ON, Canada, K7L 3N6.
Organizational Affiliation: