Salivary complement inhibitors from mosquitoes: Structure and mechanism of action.
Strayer, E.C., Lu, S., Ribeiro, J., Andersen, J.F.(2020) J Biological Chem 296: 100083-100083
- PubMed: 33199367
- DOI: https://doi.org/10.1074/jbc.RA120.015230
- Primary Citation of Related Structures:
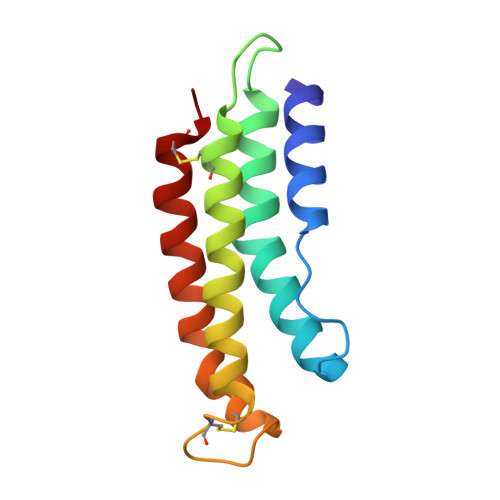
6XKE, 6XL7, 6XMB - PubMed Abstract:
Inhibition of the alternative pathway (AP) of complement by saliva from Anopheles mosquitoes facilitates feeding by blocking production of the anaphylatoxins C3a and C5a, which activate mast cells leading to plasma extravasation, pain, and itching. We have previously shown that albicin, a member of the SG7 protein family from An. Albimanus, blocks the AP by binding to and inhibiting the function of the C3 convertase, C3bBb. Here we show that SG7.AF, the albicin homolog from An. freeborni, has a similar potency to albicin but is more active in the presence of properdin, a plasma protein that acts to stabilize C3bBb. Conversely, albicin is highly active in the absence or presence of properdin. Albicin and SG7.AF stabilize the C3bBb complex in a form that accumulates on surface plasmon resonance (SPR) surfaces coated with properdin, but SG7.AF binds with lower affinity than albicin. Albicin induces oligomerization of the complex in solution, suggesting that it is oligomerization that leads to stabilization on SPR surfaces. Anophensin, the albicin ortholog from An. stephensi, is only weakly active as an inhibitor of the AP, suggesting that the SG7 family may play a different functional role in this species and other species of the subgenus Cellia, containing the major malaria vectors in Africa and Asia. Crystal structures of albicin and SG7.AF reveal a novel four-helix bundle arrangement that is stabilized by an N-terminal hydrogen bonding network. These structures provide insight into the SG7 family and related mosquito salivary proteins including the platelet-inhibitory 30 kDa family.
- Laboratory of Malaria and Vector Research, NIH-NIAID, Rockville, Maryland, USA.
Organizational Affiliation: