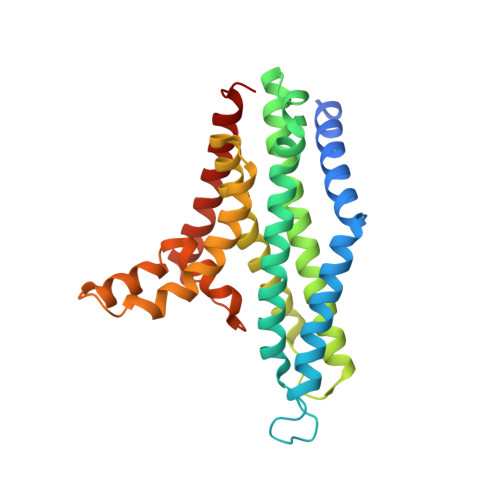
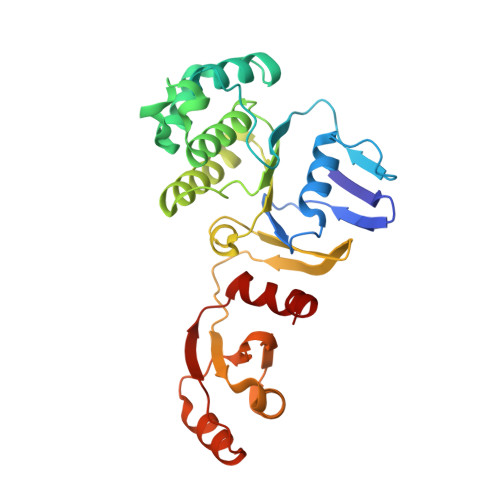
Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Zeytuni, N., Dickey, S.W., Hu, J., Chou, H.T., Worrall, L.J., Alexander, J.A.N., Carlson, M.L., Nosella, M., Duong, F., Yu, Z., Otto, M., Strynadka, N.C.J.(2020) Sci Adv 6: eabb8219
- PubMed: 32998902
- DOI: https://doi.org/10.1126/sciadv.abb8219
- Primary Citation of Related Structures:
6U2D, 6XFU, 6XJH, 6XJI - PubMed Abstract:
Staphylococcus aureus is a major human pathogen that has acquired alarming broad-spectrum antibiotic resistance. One group of secreted toxins with key roles during infection is the phenol-soluble modulins (PSMs). PSMs are amphipathic, membrane-destructive cytolytic peptides that are exported to the host-cell environment by a designated adenosine 5'-triphosphate (ATP)-binding cassette (ABC) transporter, the PSM transporter (PmtABCD). Here, we demonstrate that the minimal Pmt unit necessary for PSM export is PmtCD and provide its first atomic characterization by single-particle cryo-EM and x-ray crystallography. We have captured the transporter in the ATP-bound state at near atomic resolution, revealing a type II ABC exporter fold, with an additional cytosolic domain. Comparison to a lower-resolution nucleotide-free map displaying an "open" conformation and putative hydrophobic inner chamber of a size able to accommodate the binding of two PSM peptides provides mechanistic insight and sets the foundation for therapeutic design.
- Department of Biochemistry and Molecular Biology and the Centre for Blood Research, University of British Columbia, Vancouver, BC, Canada.
Organizational Affiliation: