The structure of PfGH50B, an agarase from the marine bacterium Pseudoalteromonas fuliginea PS47
Pluvinage, B., Robb, C.S., Jeffries, R., Boraston, A.B.(2020) Acta Crystallogr F Struct Biol Commun 76: 422-427
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2020) Acta Crystallogr F Struct Biol Commun 76: 422-427
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Agarase | 765 | Pseudoalteromonas fuliginea | Mutation(s): 0 Gene Names: EUZ79_06745 | 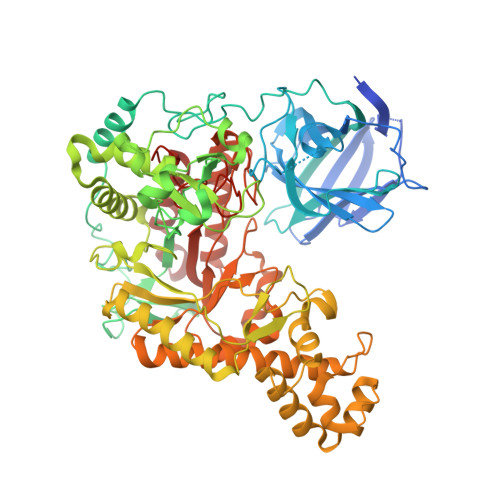 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EDO Query on EDO | C [auth A], D [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 73.24 | α = 90 |
| b = 76.26 | β = 90 |
| c = 269.86 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Natural Sciences and Engineering Research Council (NSERC, Canada) | Canada | -- |