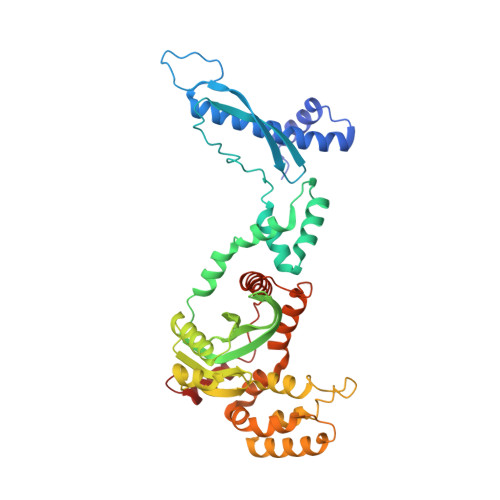
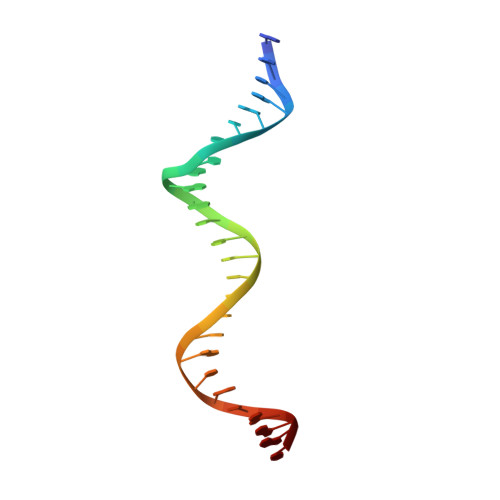
Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Kosek, D., Hickman, A.B., Ghirlando, R., He, S., Dyda, F.(2021) EMBO J 40: e105666-e105666
- PubMed: 33006208
- DOI: https://doi.org/10.15252/embj.2020105666
- Primary Citation of Related Structures:
6XG8, 6XGW, 6XGX - PubMed Abstract:
Copy-out/paste-in transposition is a major bacterial DNA mobility pathway. It contributes significantly to the emergence of antibiotic resistance, often by upregulating expression of downstream genes upon integration. Unlike other transposition pathways, it requires both asymmetric and symmetric strand transfer steps. Here, we report the first structural study of a copy-out/paste-in transposase and demonstrate its ability to catalyze all pathway steps in vitro. X-ray structures of ISCth4 transposase, a member of the IS256 family of insertion sequences, bound to DNA substrates corresponding to three sequential steps in the reaction reveal an unusual asymmetric dimeric transpososome. During transposition, an array of N-terminal domains binds a single transposon end while the catalytic domain moves to accommodate the varying substrates. These conformational changes control the path of DNA flanking the transposon end and the generation of DNA-binding sites. Our results explain the asymmetric outcome of the initial strand transfer and show how DNA binding is modulated by the asymmetric transposase to allow the capture of a second transposon end and to integrate a circular intermediate.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Organizational Affiliation: