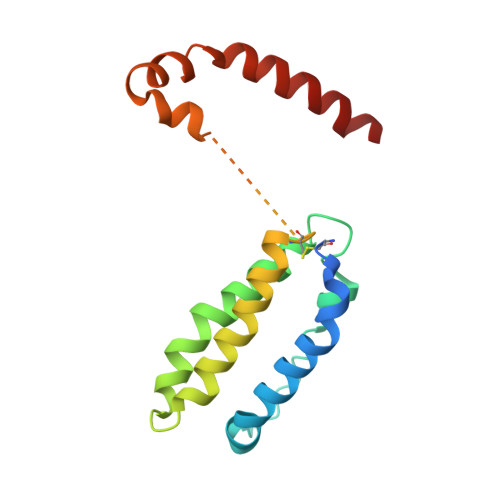
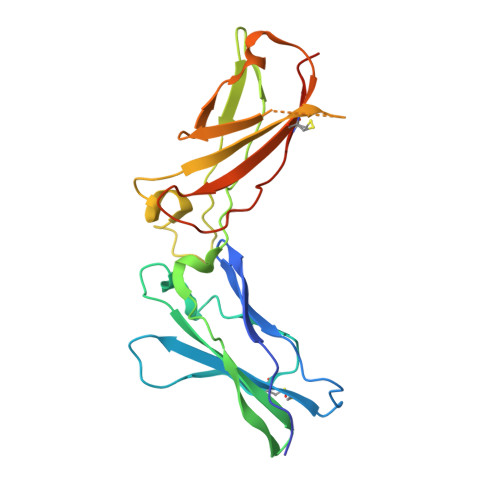
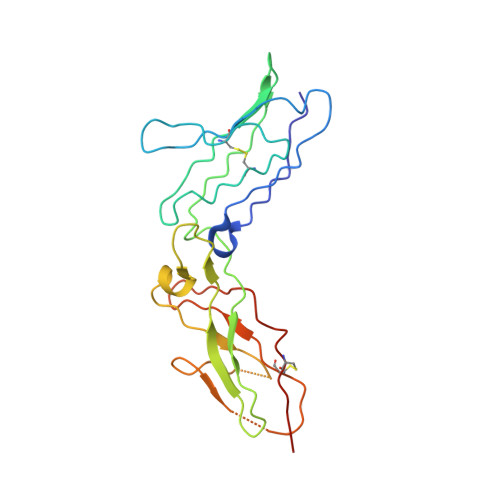
Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10.
Saxton, R.A., Tsutsumi, N., Su, L.L., Abhiraman, G.C., Mohan, K., Henneberg, L.T., Aduri, N.G., Gati, C., Garcia, K.C.(2021) Science 371
- PubMed: 33737461
- DOI: https://doi.org/10.1126/science.abc8433
- Primary Citation of Related Structures:
6X93 - PubMed Abstract:
Interleukin-10 (IL-10) is an immunoregulatory cytokine with both anti-inflammatory and immunostimulatory properties and is frequently dysregulated in disease. We used a structure-based approach to deconvolute IL-10 pleiotropy by determining the structure of the IL-10 receptor (IL-10R) complex by cryo-electron microscopy at a resolution of 3.5 angstroms. The hexameric structure shows how IL-10 and IL-10Rα form a composite surface to engage the shared signaling receptor IL-10Rβ, enabling the design of partial agonists. IL-10 variants with a range of IL-10Rβ binding strengths uncovered substantial differences in response thresholds across immune cell populations, providing a means of manipulating IL-10 cell type selectivity. Some variants displayed myeloid-biased activity by suppressing macrophage activation without stimulating inflammatory CD8 + T cells, thereby uncoupling the major opposing functions of IL-10. These results provide a mechanistic blueprint for tuning the pleiotropic actions of IL-10.
- Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: