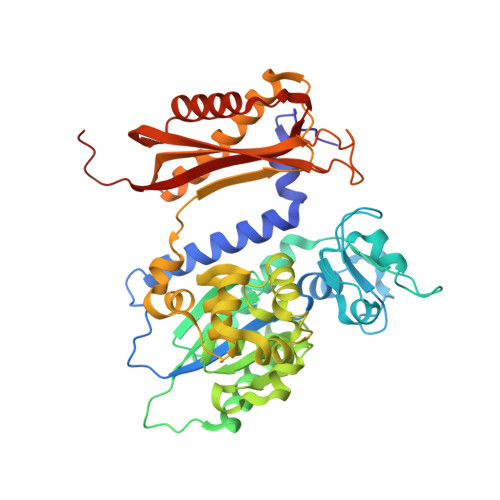
Visualizing Rev1 catalyze protein-template DNA synthesis.
Weaver, T.M., Cortez, L.M., Khoang, T.H., Washington, M.T., Agarwal, P.K., Freudenthal, B.D.(2020) Proc Natl Acad Sci U S A 117: 25494-25504
- PubMed: 32999062
- DOI: https://doi.org/10.1073/pnas.2010484117
- Primary Citation of Related Structures:
6X6Z, 6X70, 6X71, 6X72, 6X73, 6X74, 6X75, 6X76, 6X77 - PubMed Abstract:
During DNA replication, replicative DNA polymerases may encounter DNA lesions, which can stall replication forks. One way to prevent replication fork stalling is through the recruitment of specialized translesion synthesis (TLS) polymerases that have evolved to incorporate nucleotides opposite DNA lesions. Rev1 is a specialized TLS polymerase that bypasses abasic sites, as well as minor-groove and exocyclic guanine adducts. Lesion bypass is accomplished using a unique protein-template mechanism in which the templating base is evicted from the DNA helix and the incoming dCTP hydrogen bonds with an arginine side chain of Rev1. To understand the protein-template mechanism at an atomic level, we employed a combination of time-lapse X-ray crystallography, molecular dynamics simulations, and DNA enzymology on the Saccharomyces cerevisiae Rev1 protein. We find that Rev1 evicts the templating base from the DNA helix prior to binding the incoming nucleotide. Binding the incoming nucleotide changes the conformation of the DNA substrate to orient it for nucleotidyl transfer, although this is not coupled to large structural changes in Rev1 like those observed with other DNA polymerases. Moreover, we found that following nucleotide incorporation, Rev1 converts the pyrophosphate product to two monophosphates, which drives the reaction in the forward direction and prevents pyrophosphorolysis. Following nucleotide incorporation, the hydrogen bonds between the incorporated nucleotide and the arginine side chain are broken, but the templating base remains extrahelical. These postcatalytic changes prevent potentially mutagenic processive synthesis by Rev1 and facilitate dissociation of the DNA product from the enzyme.
- Department of Biochemistry and Molecular Biology, University of Kansas Medical Center, Kansas City, KS 66160.
Organizational Affiliation: