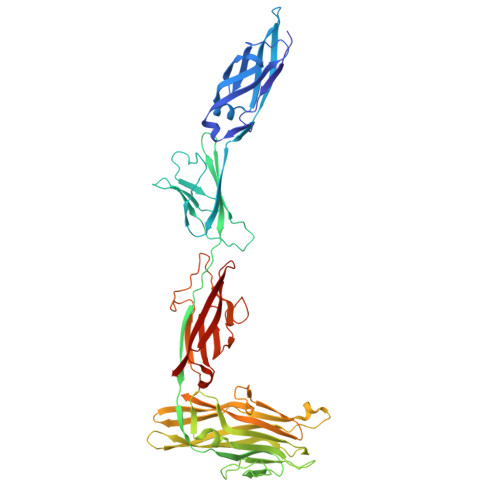
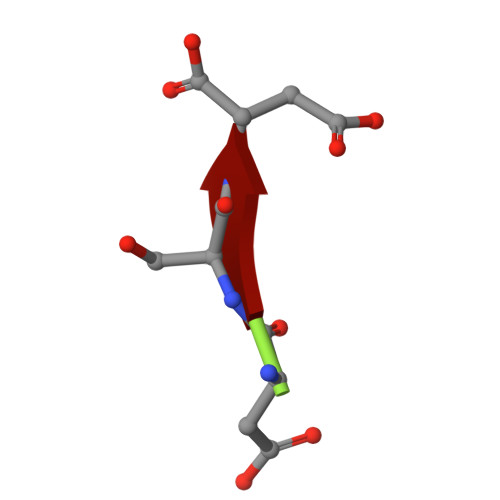
Molecular basis for inhibition of adhesin-mediated bacterial-host interactions through a peptide-binding domain.
Guo, S., Zahiri, H., Stevens, C., Spaanderman, D.C., Milroy, L.G., Ottmann, C., Brunsveld, L., Voets, I.K., Davies, P.L.(2021) Cell Rep 37: 110002-110002
- PubMed: 34788627
- DOI: https://doi.org/10.1016/j.celrep.2021.110002
- Primary Citation of Related Structures:
6X5V, 6X5W, 6X6M, 6X6Q - PubMed Abstract:
Infections typically begin with pathogens adhering to host cells. For bacteria, this adhesion can occur through specific ligand-binding domains. We identify a 20-kDa peptide-binding domain (PBD) in a 1.5-MDa RTX adhesin of a Gram-negative marine bacterium that colonizes diatoms. The crystal structure of this Ca 2+ -dependent PBD suggests that it may bind the C termini of host cell-surface proteins. A systematic peptide library analysis reveals an optimal tripeptide sequence with 30-nM affinity for the PBD, and X-ray crystallography details its peptide-protein interactions. Binding of the PBD to the diatom partner of the bacteria can be inhibited or competed away by the peptide, providing a molecular basis for inhibiting bacterium-host interactions. We further show that this PBD is found in other bacteria, including human pathogens such as Vibrio cholerae and Aeromonas veronii. Here, we produce the PBD ortholog from A. veronii and demonstrate, using the same peptide inhibitor, how pathogens may be prevented from adhering to their hosts.
- Department of Biomedical and Molecular Sciences, Queen's University, Kingston, ON K7L 3N6, Canada; Laboratory of Self-Organizing Soft Matter, Eindhoven University of Technology PO Box 513, 5600 MB Eindhoven, the Netherlands; Laboratory of Chemical Biology, Eindhoven University of Technology PO Box 513, 5600 MB Eindhoven, the Netherlands; Institute for Complex Molecular Systems, Eindhoven University of Technology PO Box 513, 5600 MB Eindhoven, the Netherlands.
Organizational Affiliation: