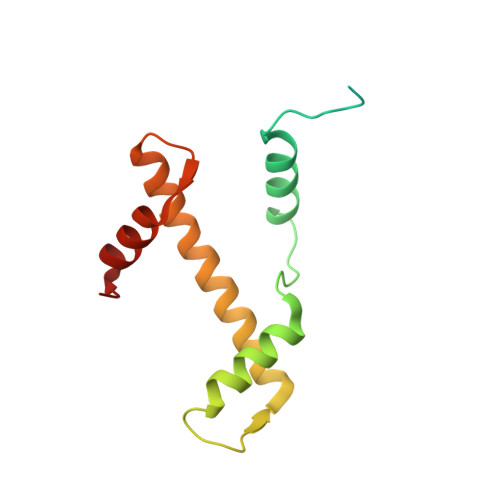
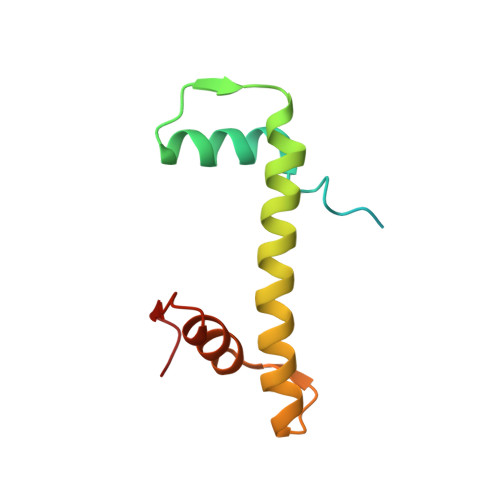
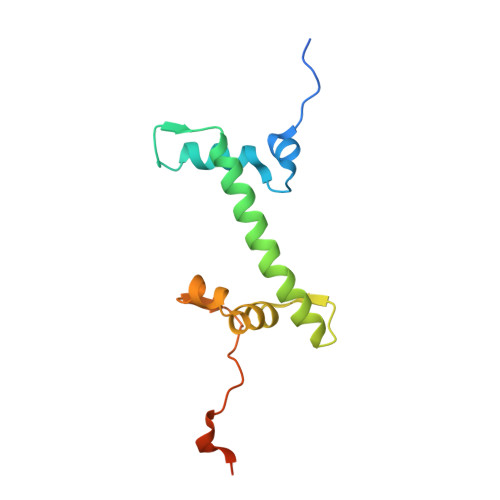
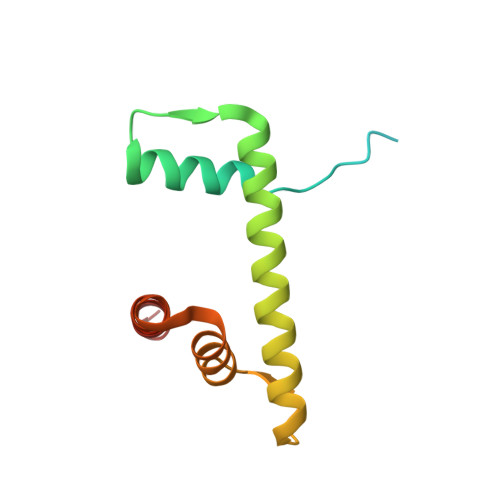
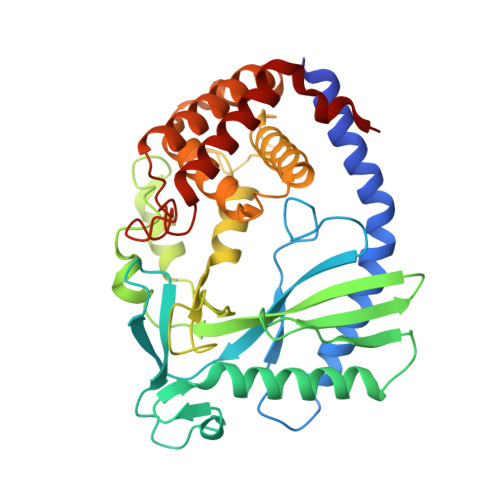
The molecular basis of tight nuclear tethering and inactivation of cGAS.
Zhao, B., Xu, P., Rowlett, C.M., Jing, T., Shinde, O., Lei, Y., West, A.P., Liu, W.R., Li, P.(2020) Nature 587: 673-677
- PubMed: 32911481
- DOI: https://doi.org/10.1038/s41586-020-2749-z
- Primary Citation of Related Structures:
6X59, 6X5A, 6XJD - PubMed Abstract:
Nucleic acids derived from pathogens induce potent innate immune responses 1-6 . Cyclic GMP-AMP synthase (cGAS) is a double-stranded DNA sensor that catalyses the synthesis of the cyclic dinucleotide cyclic GMP-AMP, which mediates the induction of type I interferons through the STING-TBK1-IRF3 signalling axis 7-11 . cGAS was previously thought to not react with self DNA owing to its cytosolic localization 2,12,13 ; however, recent studies have shown that cGAS is localized mostly in the nucleus and has low activity as a result of tight nuclear tethering 14-18 . Here we show that cGAS binds to nucleosomes with nanomolar affinity and that nucleosome binding potently inhibits its catalytic activity. To elucidate the molecular basis of cGAS inactivation by nuclear tethering, we determined the structure of mouse cGAS bound to human nucleosome by cryo-electron microscopy. The structure shows that cGAS binds to a negatively charged acidic patch formed by histones H2A and H2B via its second DNA-binding site 19 . High-affinity nucleosome binding blocks double-stranded DNA binding and maintains cGAS in an inactive conformation. Mutations of cGAS that disrupt nucleosome binding alter cGAS-mediated signalling in cells.
- Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX, USA.
Organizational Affiliation: