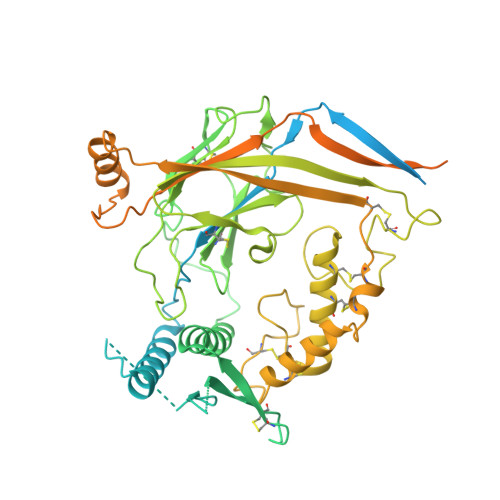
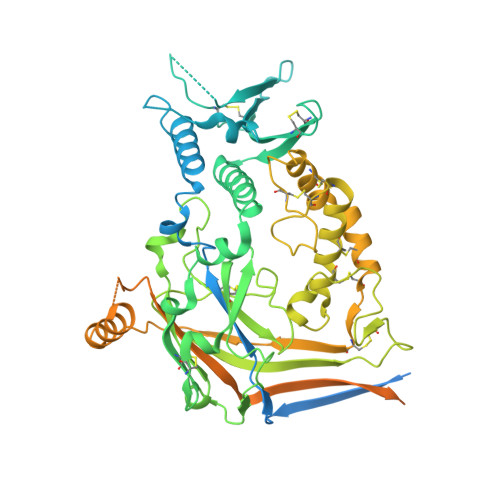
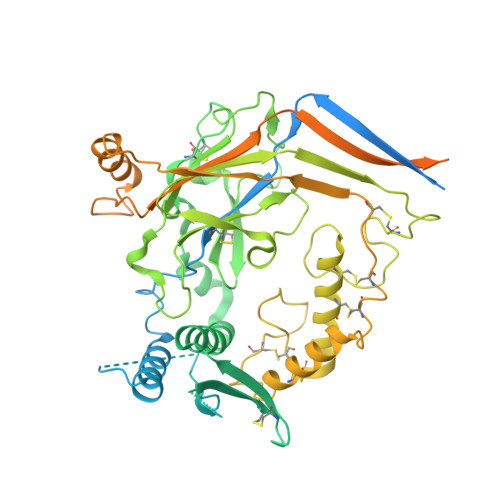
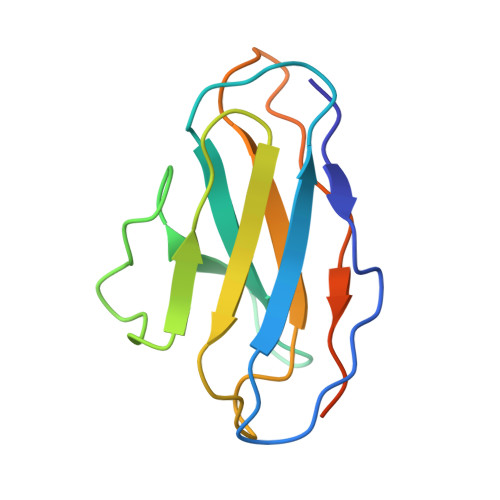
Molecular principles of assembly, activation, and inhibition in epithelial sodium channel.
Noreng, S., Posert, R., Bharadwaj, A., Houser, A., Baconguis, I.(2020) Elife 9
- PubMed: 32729833
- DOI: https://doi.org/10.7554/eLife.59038
- Primary Citation of Related Structures:
6WTH - PubMed Abstract:
The molecular bases of heteromeric assembly and link between Na + self-inhibition and protease-sensitivity in epithelial sodium channels (ENaCs) are not fully understood. Previously, we demonstrated that ENaC subunits - α, β, and γ - assemble in a counterclockwise configuration when viewed from outside the cell with the protease-sensitive GRIP domains in the periphery (Noreng et al., 2018). Here we describe the structure of ENaC resolved by cryo-electron microscopy at 3 Å. We find that a combination of precise domain arrangement and complementary hydrogen bonding network defines the subunit arrangement. Furthermore, we determined that the α subunit has a primary functional module consisting of the finger and GRIP domains. The module is bifurcated by the α2 helix dividing two distinct regulatory sites: Na + and the inhibitory peptide. Removal of the inhibitory peptide perturbs the Na + site via the α2 helix highlighting the critical role of the α2 helix in regulating ENaC function.
- Department of Biochemistry and Molecular Biology, Oregon Health & Science University, Portland, United States.
Organizational Affiliation: