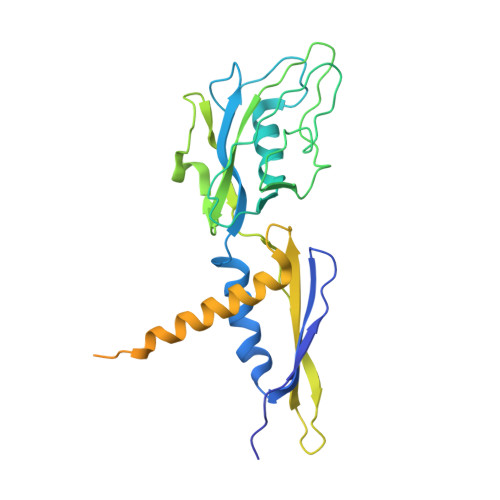
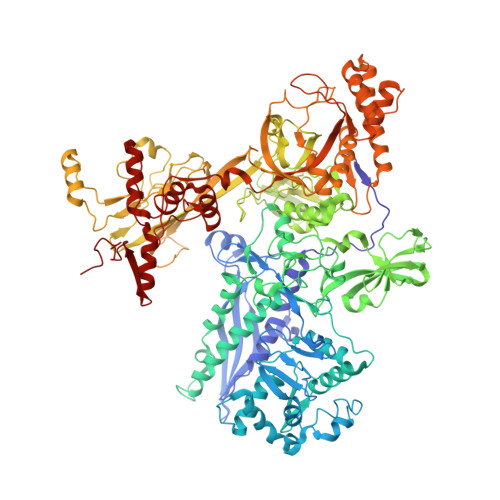
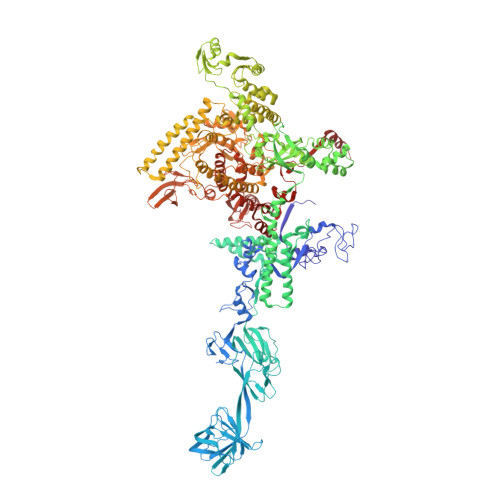
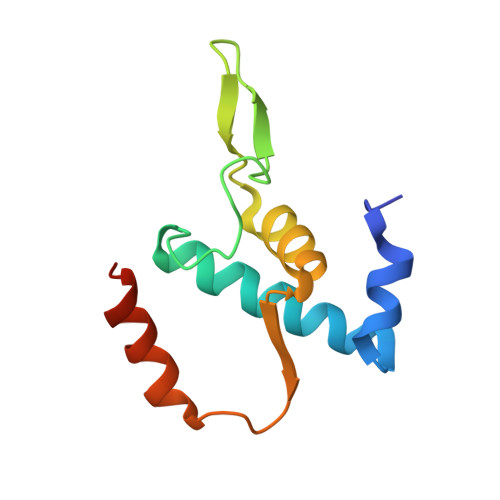
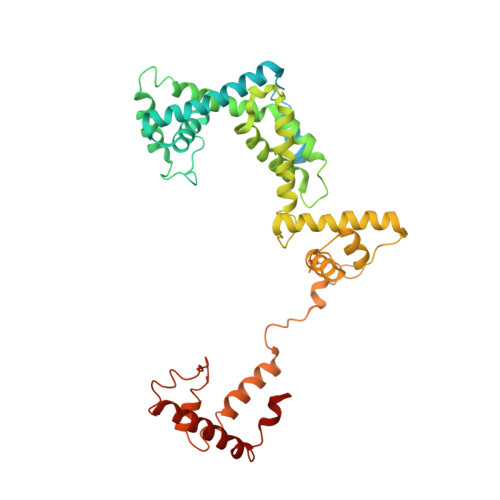
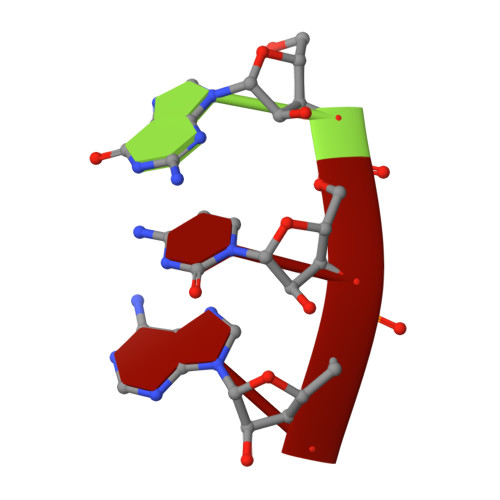
The mechanism of the nucleo-sugar selection by multi-subunit RNA polymerases.
Makinen, J.J., Shin, Y., Vieras, E., Virta, P., Metsa-Ketela, M., Murakami, K.S., Belogurov, G.A.(2021) Nat Commun 12: 796-796
- PubMed: 33542236
- DOI: https://doi.org/10.1038/s41467-021-21005-w
- Primary Citation of Related Structures:
6WOX, 6WOY - PubMed Abstract:
RNA polymerases (RNAPs) synthesize RNA from NTPs, whereas DNA polymerases synthesize DNA from 2'dNTPs. DNA polymerases select against NTPs by using steric gates to exclude the 2'OH, but RNAPs have to employ alternative selection strategies. In single-subunit RNAPs, a conserved Tyr residue discriminates against 2'dNTPs, whereas selectivity mechanisms of multi-subunit RNAPs remain hitherto unknown. Here, we show that a conserved Arg residue uses a two-pronged strategy to select against 2'dNTPs in multi-subunit RNAPs. The conserved Arg interacts with the 2'OH group to promote NTP binding, but selectively inhibits incorporation of 2'dNTPs by interacting with their 3'OH group to favor the catalytically-inert 2'-endo conformation of the deoxyribose moiety. This deformative action is an elegant example of an active selection against a substrate that is a substructure of the correct substrate. Our findings provide important insights into the evolutionary origins of biopolymers and the design of selective inhibitors of viral RNAPs.
- Department of Biochemistry, University of Turku, Turku, Finland.
Organizational Affiliation: