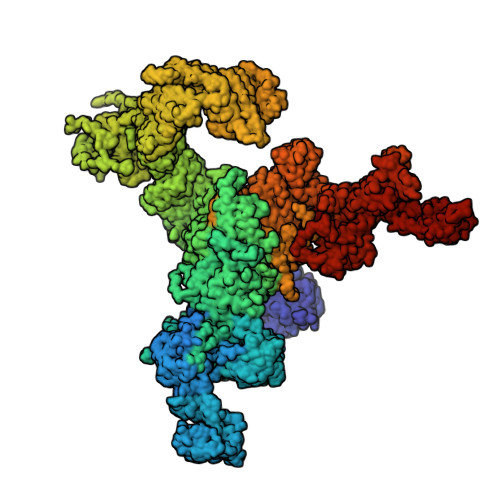
Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Iyer, K.A., Hu, Y., Nayak, A.R., Kurebayashi, N., Murayama, T., Samso, M.(2020) Sci Adv 6: eabb2964-eabb2964
- PubMed: 32832689
- DOI: https://doi.org/10.1126/sciadv.abb2964
- Primary Citation of Related Structures:
6WOT, 6WOU, 6WOV - PubMed Abstract:
Mutations in ryanodine receptors (RyRs), intracellular Ca 2+ channels, are associated with deadly disorders. Despite abundant functional studies, the molecular mechanism of RyR malfunction remains elusive. We studied two single-point mutations at an equivalent site in the skeletal (RyR1 R164C) and cardiac (RyR2 R176Q) isoforms using ryanodine binding, Ca 2+ imaging, and cryo-electron microscopy (cryo-EM) of the full-length protein. Loss of the positive charge had greater effect on the skeletal isoform, mediated via distortion of a salt bridge network, a molecular latch inducing rotation of a cytoplasmic domain, and partial progression to open-state traits of the large cytoplasmic assembly accompanied by alteration of the Ca 2+ binding site, which concur with the major "hyperactive" feature of the mutated channel. Our cryo-EM studies demonstrated the allosteric effect of a mutation situated ~85 Å away from the pore and identified an isoform-specific structural effect.
- Department of Physiology and Biophysics, School of Medicine, Virginia Commonwealth University, Richmond, VA, USA.
Organizational Affiliation: