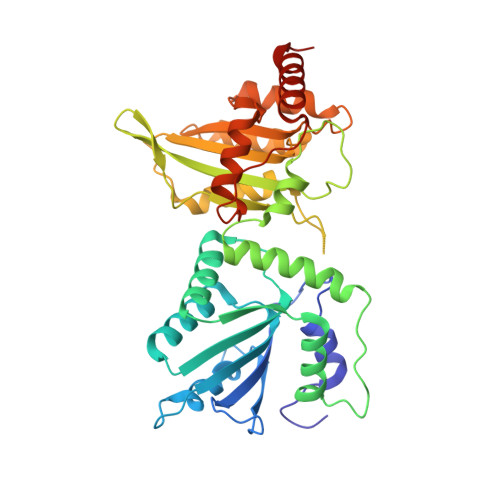
Crystal Structure of a Soluble APOBEC3G Variant Suggests ssDNA to Bind in a Channel that Extends between the Two Domains.
Maiti, A., Myint, W., Delviks-Frankenberry, K.A., Hou, S., Kanai, T., Balachandran, V., Sierra Rodriguez, C., Tripathi, R., Kurt Yilmaz, N., Pathak, V.K., Schiffer, C.A., Matsuo, H.(2020) J Mol Biology 432: 6042-6060
- PubMed: 33098858
- DOI: https://doi.org/10.1016/j.jmb.2020.10.020
- Primary Citation of Related Structures:
6WMA, 6WMB, 6WMC - PubMed Abstract:
APOBEC3G (A3G) is a single-stranded DNA (ssDNA) cytosine deaminase that can restrict HIV-1 infection by mutating the viral genome. A3G consists of a non-catalytic N-terminal domain (NTD) and a catalytic C-terminal domain (CTD) connected by a short linker. While the CTD catalyzes cytosine deamination, the NTD is believed to provide additional affinity for ssDNA. Structures of both A3G domains have been solved individually; however, a full-length A3G structure has been challenging. Recently, crystal structures of full-length rhesus macaque A3G variants were solved which suggested dimerization mechanisms and RNA binding surfaces, whereas the dimerization appeared to compromise catalytic activity. We determined the crystal structure of a soluble variant of human A3G (sA3G) at 2.5 Å and from these data generated a model structure of wild-type A3G. This model demonstrated that the NTD was rotated 90° relative to the CTD along the major axis of the molecule, an orientation that forms a positively charged channel connected to the CTD catalytic site, consisting of NTD loop-1 and CTD loop-3. Structure-based mutations, in vitro deamination and DNA binding assays, and HIV-1 restriction assays identify R24, located in the NTD loop-1, as essential to a critical interaction with ssDNA. Furthermore, sA3G was shown to bind a deoxy-cytidine dinucleotide near the catalytic Zn 2+ , yet not in the catalytic position, where the interactions between deoxy-cytidines and CTD loop-1 and loop-7 residues were different from those formed with substrate. These new interactions suggest a mechanism explaining why A3G exhibits a 3' to 5' directional preference in processive deamination.
- Basic Science Program, Frederick National Laboratory for Cancer Research, Frederick, MD 21702, USA.
Organizational Affiliation: