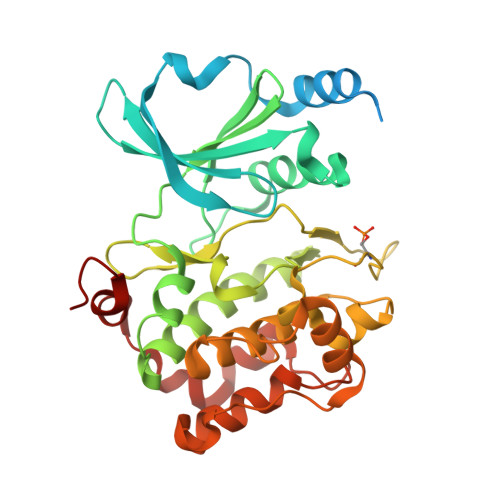
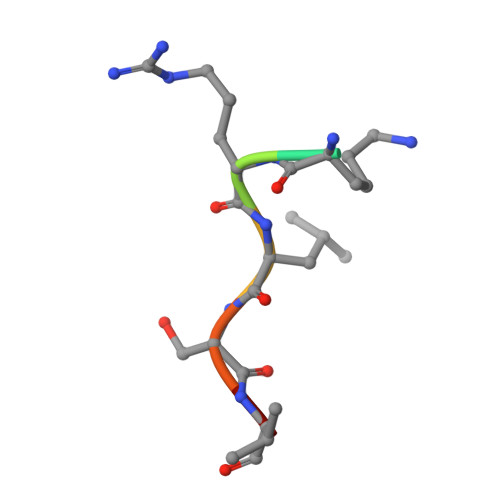
Recognition of physiological phosphorylation sites by p21-activated kinase 4.
Chetty, A.K., Sexton, J.A., Ha, B.H., Turk, B.E., Boggon, T.J.(2020) J Struct Biol 211: 107553-107553
- PubMed: 32585314
- DOI: https://doi.org/10.1016/j.jsb.2020.107553
- Primary Citation of Related Structures:
6WLX, 6WLY - PubMed Abstract:
Many serine/threonine protein kinases discriminate between serine and threonine substrates as a filter to control signaling output. Among these, the p21-activated kinase (PAK) group strongly favors phosphorylation of Ser over Thr residues. PAK4, a group II PAK, almost exclusively phosphorylates its substrates on serine residues. The only well documented exception is LIM domain kinase 1 (LIMK1), which is phosphorylated on an activation loop threonine (Thr508) to promote its catalytic activity. To understand the molecular and kinetic basis for PAK4 substrate selectivity we compared its mode of recognition of LIMK1 (Thr508) with that of a known serine substrate, β-catenin (Ser675). We determined X-ray crystal structures of PAK4 in complex with synthetic peptides corresponding to its phosphorylation sites in LIMK1 and β-catenin to 1.9 Å and 2.2 Å resolution, respectively. We found that the PAK4 DFG + 1 residue, a key determinant of phosphoacceptor preference, adopts a sub-optimal orientation when bound to LIMK1 compared to β-catenin. In peptide kinase activity assays, we find that phosphoacceptor identity impacts catalytic efficiency but does not affect the K m value for both phosphorylation sites. Although catalytic efficiency of wild-type LIMK1 and β-catenin are equivalent, T508S mutation of LIMK1 creates a highly efficient substrate. These results suggest suboptimal phosphorylation of LIMK1 as a mechanism for controlling the dynamics of substrate phosphorylation by PAK4.
- Yale College, New Haven, CT 06520, USA; Department of Molecular Biophysics and Biochemistry, Yale University, 333 Cedar Street, New Haven, CT 06520, USA.
Organizational Affiliation: