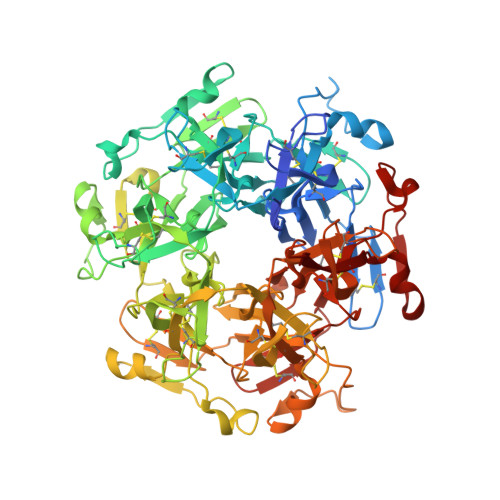
Collagen IV alpha 345 dysfunction in glomerular basement membrane diseases. II. Crystal structure of the alpha 345 hexamer.
Boudko, S.P., Bauer, R., Chetyrkin, S.V., Ivanov, S., Smith, J., Voziyan, P.A., Hudson, B.G.(2021) J Biological Chem 296: 100591-100591
- PubMed: 33775698
- DOI: https://doi.org/10.1016/j.jbc.2021.100591
- Primary Citation of Related Structures:
6WKU - PubMed Abstract:
Our recent work identified a genetic variant of the α345 hexamer of the collagen IV scaffold that is present in patients with glomerular basement membrane diseases, Goodpasture's disease (GP) and Alport syndrome (AS), and phenocopies of AS in knock-in mice. To understand the context of this "Zurich" variant, an 8-amino acid appendage, we developed a construct of the WT α345 hexamer using the single-chain NC1 trimer technology, which allowed us to solve a crystal structure of this key connection module. The α345 hexamer structure revealed a ring of 12 chloride ions at the trimer-trimer interface, analogous to the collagen α121 hexamer, and the location of the 170 AS variants. The hexamer surface is marked by multiple pores and crevices that are potentially accessible to small molecules. Loop-crevice-loop features constitute bioactive sites, where pathogenic pathways converge that are linked to AS and GP, and, potentially, diabetic nephropathy. In Pedchenko et al., we demonstrate that these sites exhibit conformational plasticity, a dynamic property underlying assembly of bioactive sites and hexamer dysfunction. The α345 hexamer structure is a platform to decipher how variants cause AS and how hypoepitopes can be triggered, causing GP. Furthermore, the bioactive sites, along with the pores and crevices on the hexamer surface, are prospective targets for therapeutic interventions.
- Department of Medicine, Division of Nephrology and Hypertension, Vanderbilt University Medical Center, Nashville, Tennessee, USA; Center for Matrix Biology, Vanderbilt University Medical Center, Nashville, Tennessee, USA; Department of Biochemistry, Center for Structural Biology, Vanderbilt University, Nashville, Tennessee, USA. Electronic address: Sergey.Budko@vumc.org.
Organizational Affiliation: