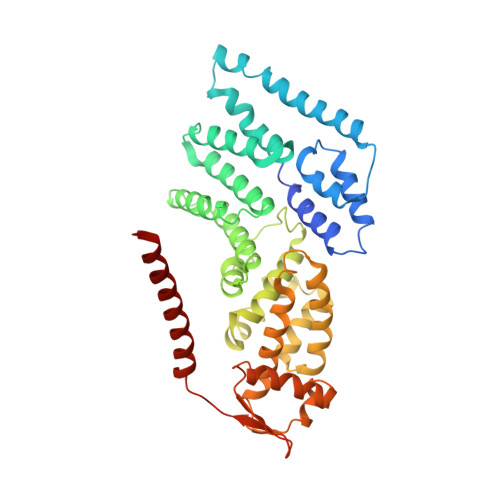
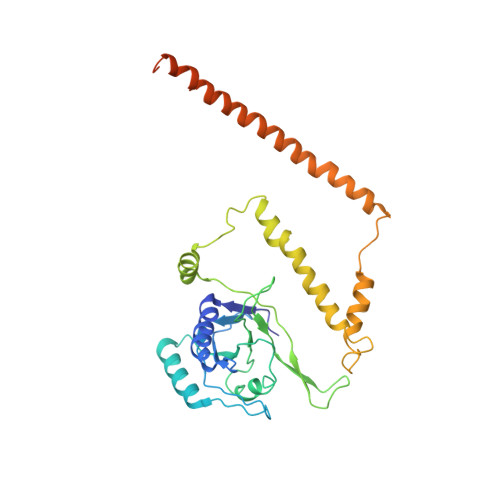
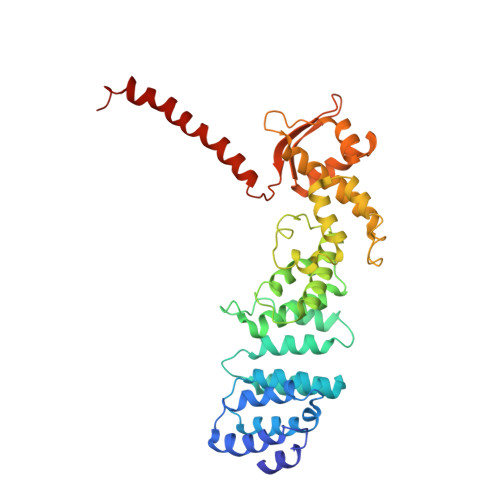
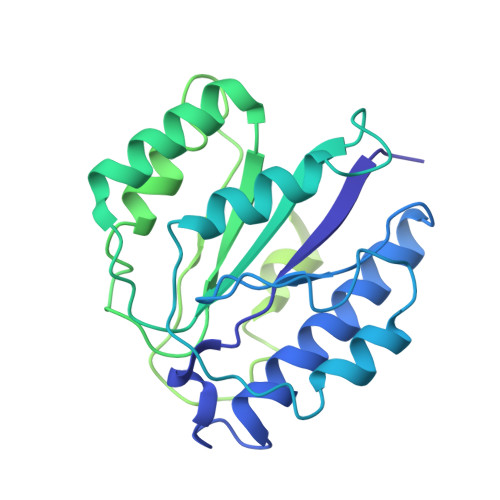
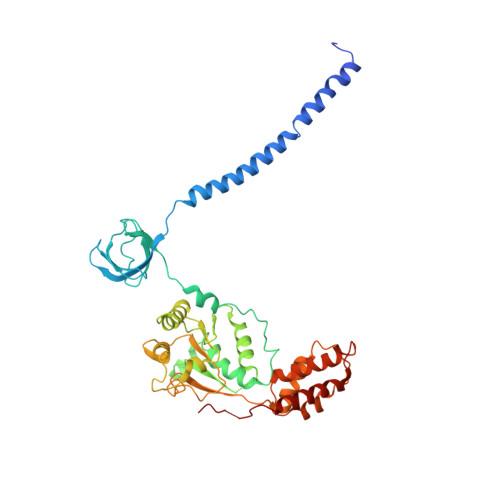
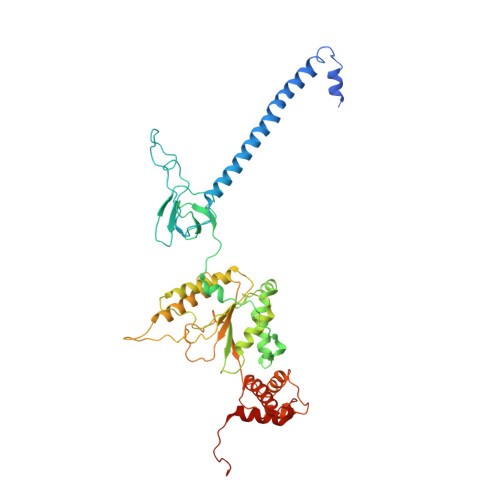
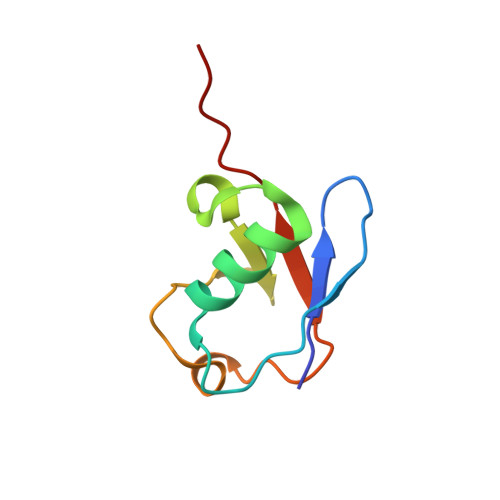
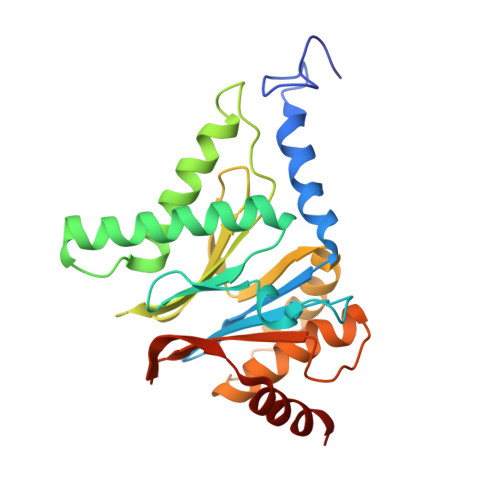
Cryo-EM Reveals Unanchored M1-Ubiquitin Chain Binding at hRpn11 of the 26S Proteasome.
Chen, X., Dorris, Z., Shi, D., Huang, R.K., Khant, H., Fox, T., de Val, N., Williams, D., Zhang, P., Walters, K.J.(2020) Structure 28: 1206-1217.e4
- PubMed: 32783951
- DOI: https://doi.org/10.1016/j.str.2020.07.011
- Primary Citation of Related Structures:
6WJD, 6WJN - PubMed Abstract:
The 26S proteasome is specialized for regulated protein degradation and formed by a dynamic regulatory particle (RP) that caps a hollow cylindrical core particle (CP) where substrates are proteolyzed. Its diverse substrates unify as proteasome targets by ubiquitination. We used cryogenic electron microscopy (cryo-EM) to study how human 26S proteasome interacts with M1-linked hexaubiquitin (M1-Ub 6 ) unanchored to a substrate and E3 ubiquitin ligase E6AP/UBE3A. Proteasome structures are available with model substrates extending through the RP ATPase ring and substrate-conjugated K63-linked ubiquitin chains present at inhibited deubiquitinating enzyme hRpn11 and the nearby ATPase hRpt4/hRpt5 coiled coil. In this study, we find M1-Ub 6 at the hRpn11 site despite the absence of conjugated substrate, indicating that ubiquitin binding at this location does not require substrate interaction with the RP. Moreover, unanchored M1-Ub 6 binds to this hRpn11 site of the proteasome with the CP gating residues in both the closed and opened conformational states.
- Protein Processing Section, Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.
Organizational Affiliation: