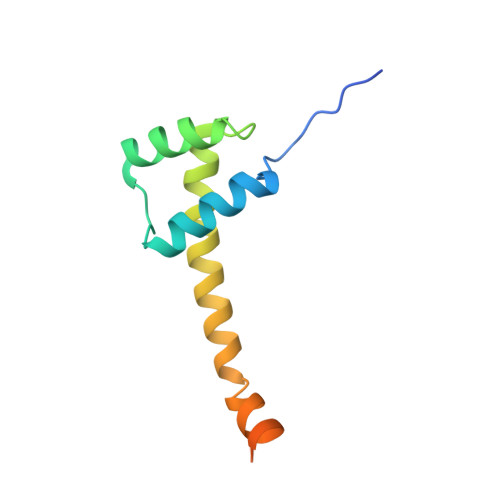
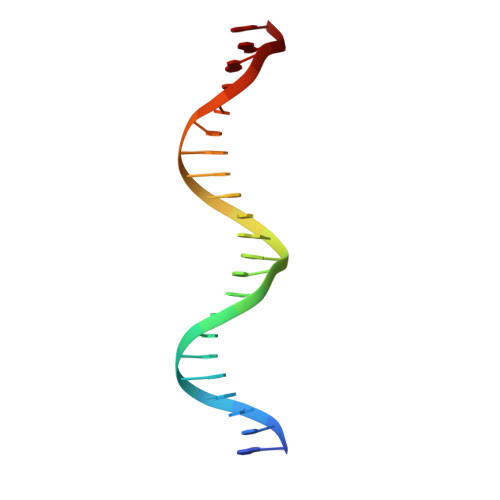
Structure of the unique tetrameric STENOFOLIA homeodomain bound with target promoter DNA.
Pathak, P.K., Zhang, F., Peng, S., Niu, L., Chaturvedi, J., Elliott, J., Xiang, Y., Tadege, M., Deng, J.(2021) Acta Crystallogr D Struct Biol 77: 1050-1063
- PubMed: 34342278
- DOI: https://doi.org/10.1107/S205979832100632X
- Primary Citation of Related Structures:
6WIG - PubMed Abstract:
Homeobox transcription factors are key regulators of morphogenesis and development in both animals and plants. In plants, the WUSCHEL-related homeobox (WOX) family of transcription factors function as central organizers of several developmental programs ranging from embryo patterning to meristematic stem-cell maintenance through transcriptional activation and repression mechanisms. The Medicago truncatula STENOFOLIA (STF) gene is a master regulator of leaf-blade lateral development. Here, the crystal structure of the homeodomain (HD) of STF (STF-HD) in complex with its promoter DNA is reported at 2.1 Å resolution. STF-HD binds DNA as a tetramer, enclosing nearly the entire bound DNA surface. The STF-HD tetramer is partially stabilized by docking of the C-terminal tail of one protomer onto a conserved hydrophobic surface on the head of another protomer in a head-to-tail manner. STF-HD specifically binds TGA motifs, although the promoter sequence also contains TAAT motifs. Helix α3 not only serves a canonical role as a base reader in the major groove, but also provides DNA binding in the minor groove through basic residues located at its C-terminus. The structural and functional data in planta reported here provide new insights into the DNA-binding mechanisms of plant-specific HDs from the WOX family of transcription factors.
- Department of Biochemistry and Molecular Biology, Oklahoma State University, Stillwater, OK 74078, USA.
Organizational Affiliation: