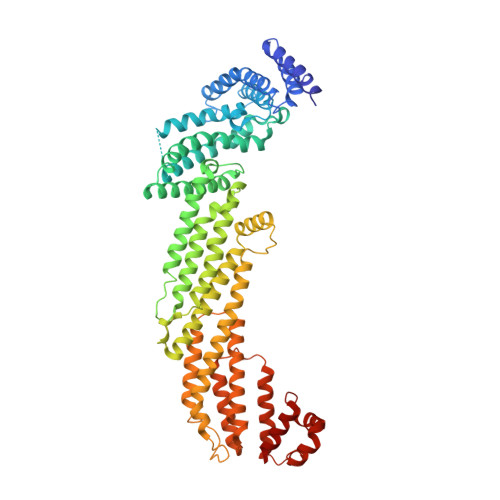
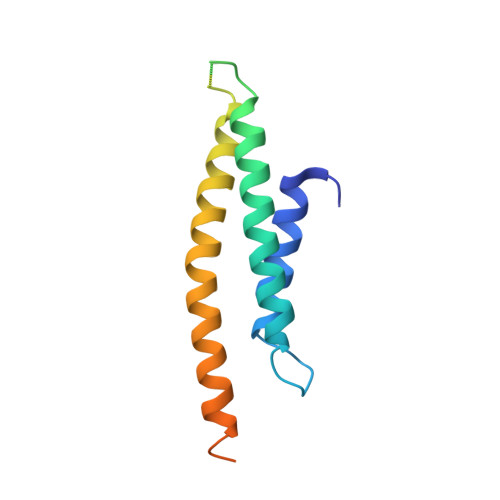
Structural basis for the binding of SNAREs to the multisubunit tethering complex Dsl1.
Travis, S.M., DAmico, K., Yu, I.M., McMahon, C., Hamid, S., Ramirez-Arellano, G., Jeffrey, P.D., Hughson, F.M.(2020) J Biological Chem 295: 10125-10135
- PubMed: 32409579
- DOI: https://doi.org/10.1074/jbc.RA120.013654
- Primary Citation of Related Structures:
6WC3 - PubMed Abstract:
Multisubunit-tethering complexes (MTCs) are large (250 to >750 kDa), conserved macromolecular machines that are essential for soluble N -ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-mediated membrane fusion in all eukaryotes. MTCs are thought to organize membrane trafficking by mediating the initial long-range interaction between a vesicle and its target membrane and promoting the formation of membrane-bridging SNARE complexes. Previously, we reported the structure of the yeast Dsl1 complex, the simplest known MTC, which is essential for coat protein I (COPI) mediated transport from the Golgi to the endoplasmic reticulum (ER). This structure suggests how the Dsl1 complex might tether a vesicle to its target membrane by binding at one end to the COPI coat and at the other to ER-associated SNAREs. Here, we used X-ray crystallography to investigate these Dsl1-SNARE interactions in greater detail. The Dsl1 complex comprises three subunits that together form a two-legged structure with a central hinge. We found that distal regions of each leg bind N-terminal Habc domains of the ER SNAREs Sec20 (a Qb-SNARE) and Use1 (a Qc-SNARE). The observed binding modes appear to anchor the Dsl1 complex to the ER target membrane while simultaneously ensuring that both SNAREs are in open conformations, with their SNARE motifs available for assembly. The proximity of the two SNARE motifs, and therefore their ability to enter the same SNARE complex, will depend on the relative orientation of the two Dsl1 legs. These results underscore the critical roles of SNARE N-terminal domains in mediating interactions with other elements of the vesicle docking and fusion machinery.
- Department of Molecular Biology, Princeton University, Princeton, New Jersey, USA.
Organizational Affiliation: