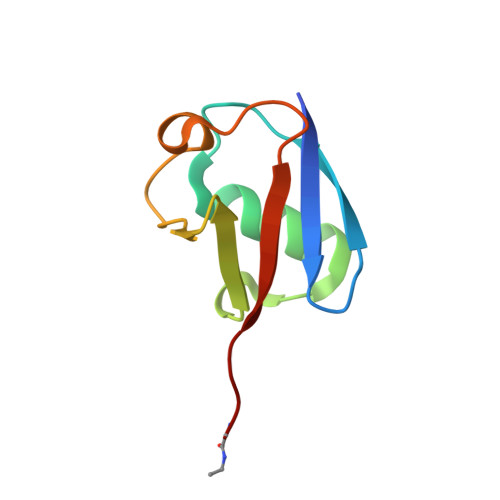
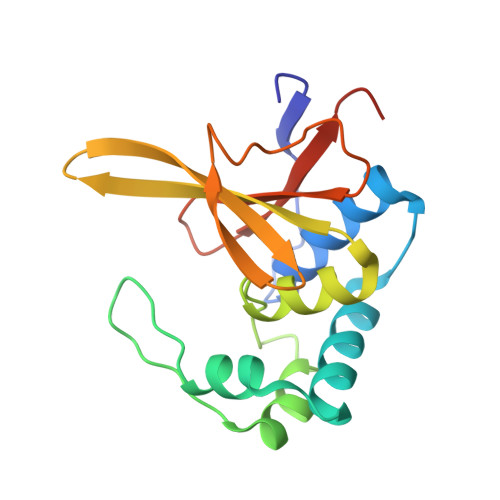
Identification and characterization of diverse OTU deubiquitinases in bacteria.
Schubert, A.F., Nguyen, J.V., Franklin, T.G., Geurink, P.P., Roberts, C.G., Sanderson, D.J., Miller, L.N., Ovaa, H., Hofmann, K., Pruneda, J.N., Komander, D.(2020) EMBO J 39: e105127-e105127
- PubMed: 32567101
- DOI: https://doi.org/10.15252/embj.2020105127
- Primary Citation of Related Structures:
6W9O, 6W9R, 6W9S - PubMed Abstract:
Manipulation of host ubiquitin signaling is becoming an increasingly apparent evolutionary strategy among bacterial and viral pathogens. By removing host ubiquitin signals, for example, invading pathogens can inactivate immune response pathways and evade detection. The ovarian tumor (OTU) family of deubiquitinases regulates diverse ubiquitin signals in humans. Viral pathogens have also extensively co-opted the OTU fold to subvert host signaling, but the extent to which bacteria utilize the OTU fold was unknown. We have predicted and validated a set of OTU deubiquitinases encoded by several classes of pathogenic bacteria. Biochemical assays highlight the ubiquitin and polyubiquitin linkage specificities of these bacterial deubiquitinases. By determining the ubiquitin-bound structures of two examples, we demonstrate the novel strategies that have evolved to both thread an OTU fold and recognize a ubiquitin substrate. With these new examples, we perform the first cross-kingdom structural analysis of the OTU fold that highlights commonalities among distantly related OTU deubiquitinases.
- Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: