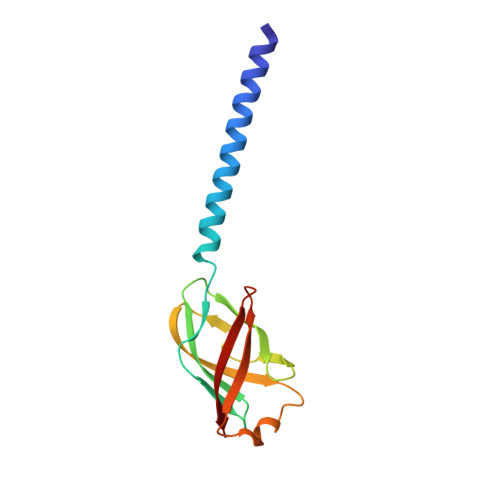
The structures of two archaeal type IV pili illuminate evolutionary relationships.
Wang, F., Baquero, D.P., Su, Z., Beltran, L.C., Prangishvili, D., Krupovic, M., Egelman, E.H.(2020) Nat Commun 11: 3424-3424
- PubMed: 32647180
- DOI: https://doi.org/10.1038/s41467-020-17268-4
- Primary Citation of Related Structures:
6W8U, 6W8X - PubMed Abstract:
We have determined the cryo-electron microscopic (cryo-EM) structures of two archaeal type IV pili (T4P), from Pyrobaculum arsenaticum and Saccharolobus solfataricus, at 3.8 Å and 3.4 Å resolution, respectively. This triples the number of high resolution archaeal T4P structures, and allows us to pinpoint the evolutionary divergence of bacterial T4P, archaeal T4P and archaeal flagellar filaments. We suggest that extensive glycosylation previously observed in T4P of Sulfolobus islandicus is a response to an acidic environment, as at even higher temperatures in a neutral environment much less glycosylation is present for Pyrobaculum than for Sulfolobus and Saccharolobus pili. Consequently, the Pyrobaculum filaments do not display the remarkable stability of the Sulfolobus filaments in vitro. We identify the Saccharolobus and Pyrobaculum T4P as host receptors recognized by rudivirus SSRV1 and tristromavirus PFV2, respectively. Our results illuminate the evolutionary relationships among bacterial and archaeal T4P filaments and provide insights into archaeal virus-host interactions.
- Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA, 22908, USA.
Organizational Affiliation: