DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Adam, S., Anteneh, H., Hornisch, M., Wagner, V., Lu, J., Radde, N.E., Bashtrykov, P., Song, J., Jeltsch, A.(2020) Nat Commun
Experimental Data Snapshot
Starting Model: experimental
View more details
(2020) Nat Commun
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA (cytosine-5)-methyltransferase 1 | 873 | Mus musculus | Mutation(s): 0 Gene Names: Dnmt1, Dnmt, Met1, Uim EC: 2.1.1.37 | 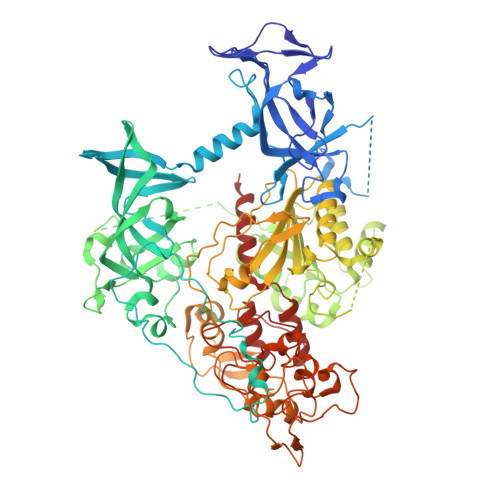 | |
UniProt | |||||
Find proteins for P13864 (Mus musculus) Explore P13864 Go to UniProtKB: P13864 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13864 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| CCG DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3') | C, D [auth E] | 12 | synthetic construct | 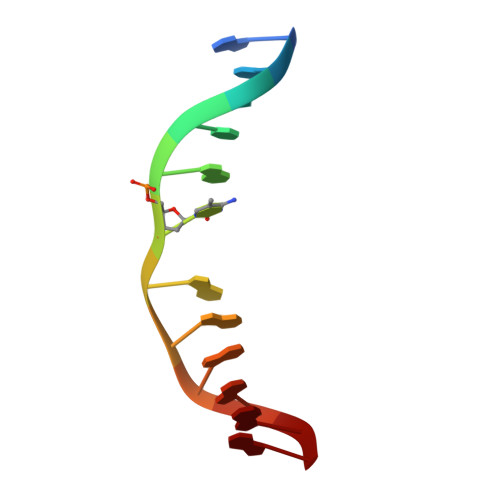 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| CCG DNA (5'-D(*CP*CP*TP*TP*CP*(C49)P*GP*TP*AP*AP*GP*T)-3') | E [auth D], F | 12 | synthetic construct | 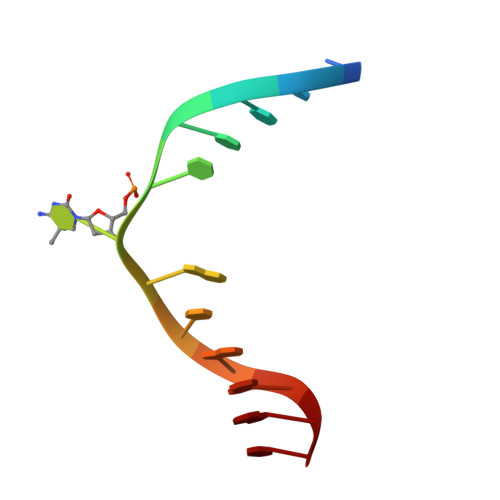 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SAH (Subject of Investigation/LOI) Query on SAH | G [auth A], J [auth B] | S-ADENOSYL-L-HOMOCYSTEINE C14 H20 N6 O5 S ZJUKTBDSGOFHSH-WFMPWKQPSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | H [auth A], I [auth A], K [auth B], L [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 89.794 | α = 90 |
| b = 152.593 | β = 94.74 |
| c = 95.631 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 1R35GM119721 |