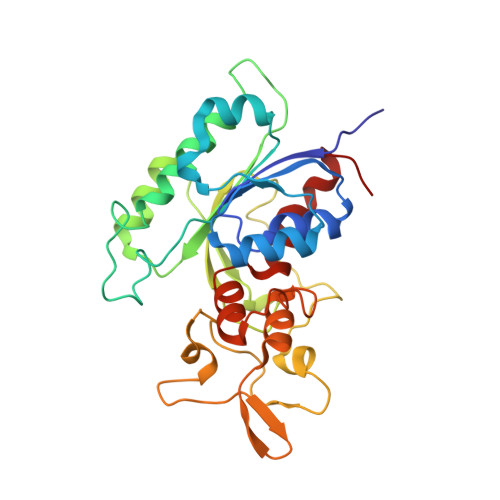
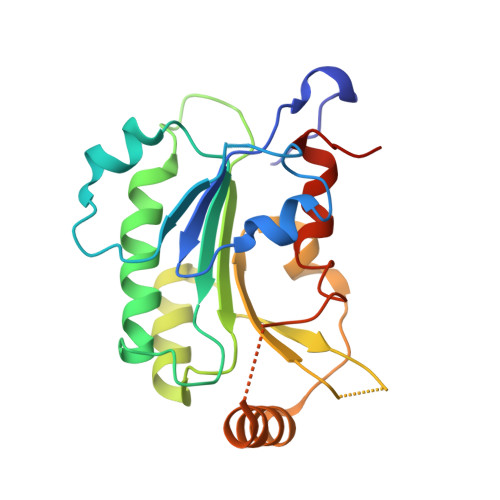
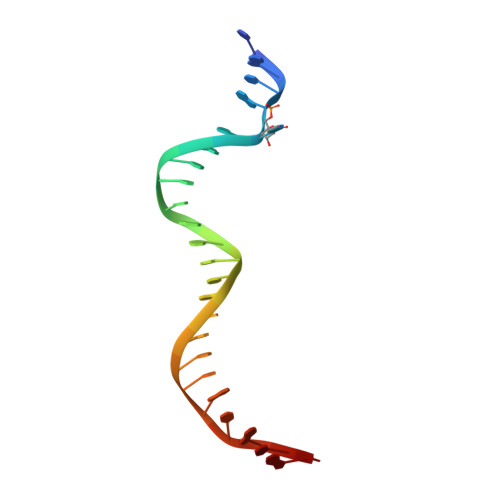
Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Anteneh, H., Fang, J., Song, J.(2020) Nat Commun 11: 2294-2294
- PubMed: 32385248
- DOI: https://doi.org/10.1038/s41467-020-16213-9
- Primary Citation of Related Structures:
6W89, 6W8B, 6W8D, 6W8J - PubMed Abstract:
DNA methyltransferase DNMT3A is essential for establishment of mammalian DNA methylation during development. The R882H DNMT3A is a hotspot mutation in acute myeloid leukemia (AML) causing aberrant DNA methylation. However, how this mutation affects the structure and function of DNMT3A remains unclear. Here we report structural characterization of wild-type and R882H-mutated DNMT3A in complex with DNA substrates with different sequence contexts. A loop from the target recognition domain (TRD loop) recognizes the CpG dinucleotides in a +1 flanking site-dependent manner. The R882H mutation reduces the DNA binding at the homodimeric interface, as well as the molecular link between the homodimeric interface and TRD loop, leading to enhanced dynamics of TRD loop. Consistently, in vitro methylation analyses indicate that the R882H mutation compromises the enzymatic activity, CpG specificity and flanking sequence preference of DNMT3A. Together, this study uncovers multiple defects of DNMT3A caused by the R882H mutation in AML.
- Department of Biochemistry, University of California, Riverside, CA, 92521, USA.
Organizational Affiliation: