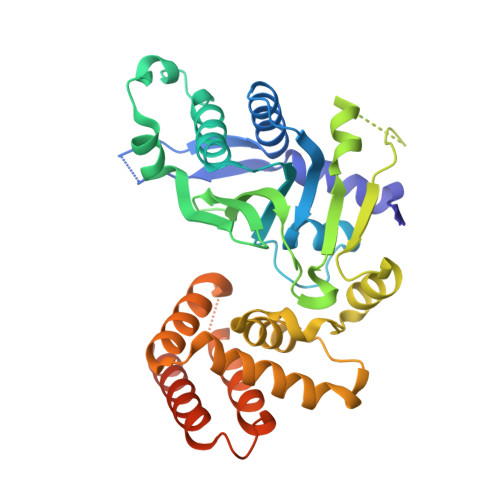
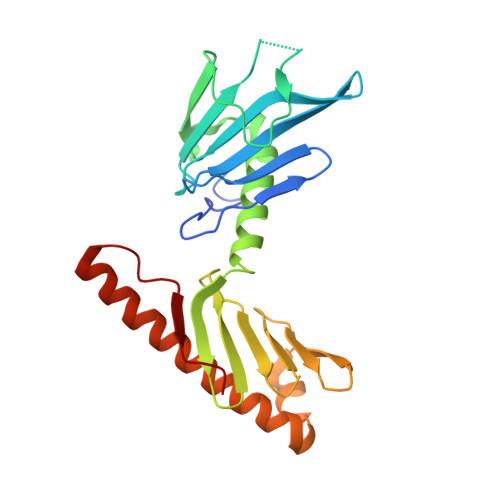
Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Chen, H., Lu, D., Shang, G., Gao, G., Zhang, X.(2020) Structure 28: 910-921.e4
- PubMed: 32433990
- DOI: https://doi.org/10.1016/j.str.2020.04.023
- Primary Citation of Related Structures:
6W36, 6W38, 6W3I, 6W3J - PubMed Abstract:
FAM46C, a non-canonical poly(A) polymerase, is frequently mutated in multiple myeloma. Loss of function of FAM46C promotes cell survival of multiple myeloma, suggesting a tumor-suppressive role. FAM46C is also essential for fastening sperm head and flagellum, indispensable for male fertility. The molecular mechanisms of these functions of FAM46C remain elusive. We report the crystal structure of FAM46C to provide the basis for its poly(A) polymerase activity and rationalize mutations associated with multiple myeloma. In addition, we found that FAM46C interacts directly with the serine/threonine kinase Plk4, the master regulator of centrosome duplication. We present the structure of FAM46C in complex with the Cryptic Polo-Box 1-2 domains of Plk4. Our structure-based mutational analyses show that the interaction with Plk4 recruits FAM46C to centrosomes. Our data suggest that Plk4-mediated localization of FAM46C enables its regulation of centrosome structure and functions, which may underlie the roles for FAM46C in cell proliferation and sperm development.
- Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: