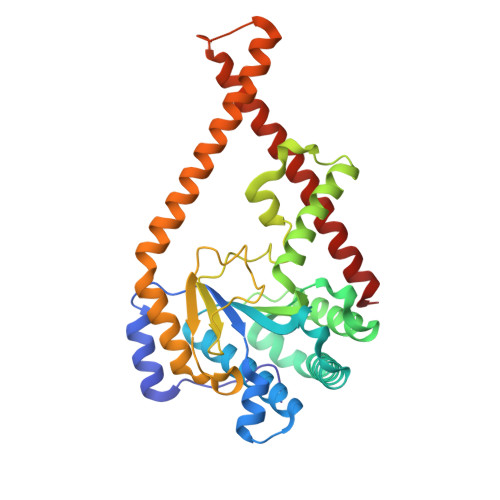
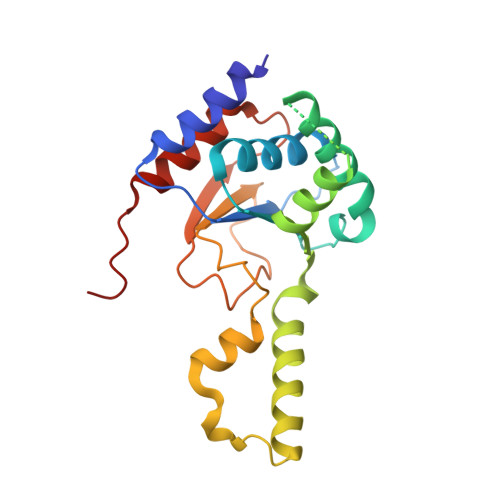
Structural elucidation of thecis-prenyltransferase NgBR/DHDDS complex reveals insights in regulation of protein glycosylation.
Edani, B.H., Grabinska, K.A., Zhang, R., Park, E.J., Siciliano, B., Surmacz, L., Ha, Y., Sessa, W.C.(2020) Proc Natl Acad Sci U S A 117: 20794-20802
- PubMed: 32817466
- DOI: https://doi.org/10.1073/pnas.2008381117
- Primary Citation of Related Structures:
6W2L - PubMed Abstract:
Cis- prenyltransferase ( cis- PTase) catalyzes the rate-limiting step in the synthesis of glycosyl carrier lipids required for protein glycosylation in the lumen of endoplasmic reticulum. Here, we report the crystal structure of the human NgBR/DHDDS complex, which represents an atomic resolution structure for any heterodimeric cis -PTase. The crystal structure sheds light on how NgBR stabilizes DHDDS through dimerization, participates in the enzyme's active site through its C-terminal -RXG- motif, and how phospholipids markedly stimulate cis -PTase activity. Comparison of NgBR/DHDDS with homodimeric cis -PTase structures leads to a model where the elongating isoprene chain extends beyond the enzyme's active site tunnel, and an insert within the α3 helix helps to stabilize this energetically unfavorable state to enable long-chain synthesis to occur. These data provide unique insights into how heterodimeric cis -PTases have evolved from their ancestral, homodimeric forms to fulfill their function in long-chain polyprenol synthesis.
- Vascular Biology and Therapeutics Program, Yale University School of Medicine, New Haven, CT 06520.
Organizational Affiliation: