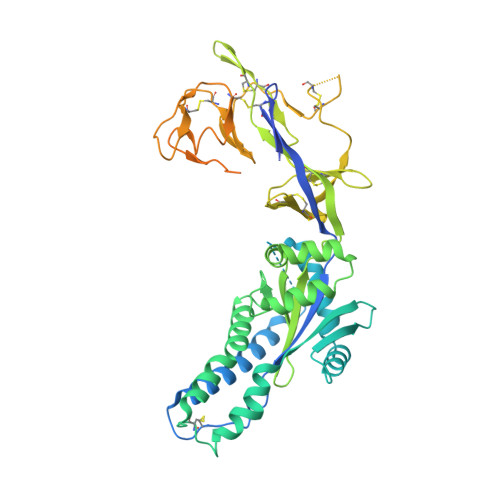
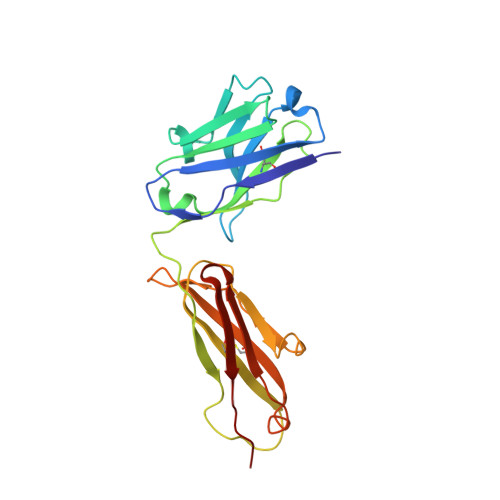
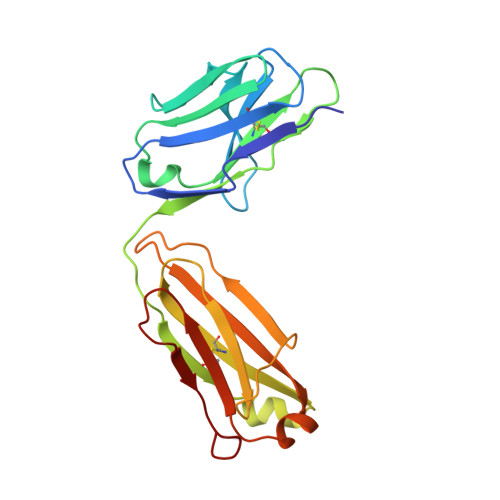
Antibody recognition of the Pneumovirus fusion protein trimer interface.
Huang, J., Diaz, D., Mousa, J.J.(2020) PLoS Pathog 16: e1008942-e1008942
- PubMed: 33035266
- DOI: https://doi.org/10.1371/journal.ppat.1008942
- Primary Citation of Related Structures:
6W16 - PubMed Abstract:
Human metapneumovirus (hMPV) is a leading cause of viral respiratory infection in children, and can cause severe lower respiratory tract infection in infants, the elderly, and immunocompromised patients. However, there remain no licensed vaccines or specific treatments for hMPV infection. Although the hMPV fusion (F) protein is the sole target of neutralizing antibodies, the immunological properties of hMPV F remain poorly understood. To further define the humoral immune response to the hMPV F protein, we isolated two new human monoclonal antibodies (mAbs), MPV458 and MPV465. Both mAbs are neutralizing in vitro and were determined to target a unique antigenic site using competitive biolayer interferometry. We determined both MPV458 and MPV465 have higher affinity for monomeric hMPV F than trimeric hMPV F. MPV458 was co-crystallized with hMPV F, and the mAb primarily interacts with an alpha helix on the F2 region of the hMPV F protein. Surprisingly, the major epitope for MPV458 lies within the trimeric interface of the hMPV F protein, suggesting significant breathing of the hMPV F protein must occur for host immune recognition of the novel epitope. In addition, significant glycan interactions were observed with a somatically mutated light chain framework residue. The data presented identifies a novel epitope on the hMPV F protein for epitope-based vaccine design, and illustrates a new mechanism for human antibody neutralization of viral glycoproteins.
- Department of Infectious Diseases, College of Veterinary Medicine, University of Georgia, Athens, GA, United States of America.
Organizational Affiliation: