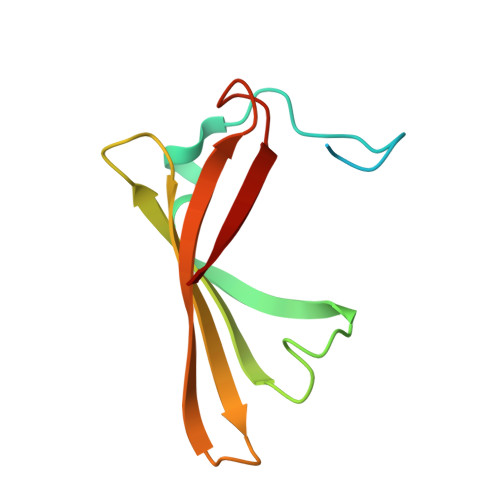
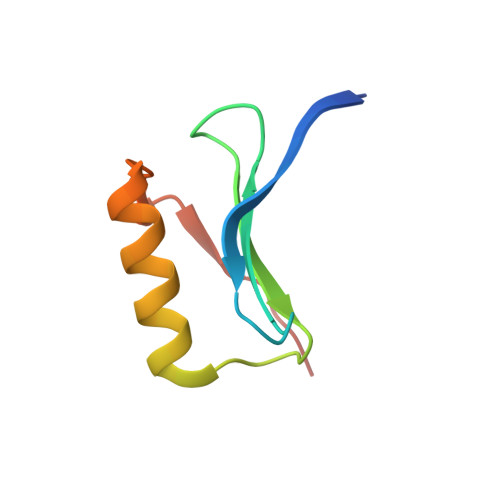
Lipocalin Blc is a potential heme-binding protein.
Bozhanova, N.G., Calcutt, M.W., Beavers, W.N., Brown, B.P., Skaar, E.P., Meiler, J.(2021) FEBS Lett 595: 206-219
- PubMed: 33210733
- DOI: https://doi.org/10.1002/1873-3468.14001
- Primary Citation of Related Structures:
6VRI - PubMed Abstract:
Lipocalins are a superfamily of functionally diverse proteins defined by a well-conserved tertiary structure despite variation in sequence. Lipocalins bind and transport small hydrophobic molecules in organisms of all kingdoms. However, there is still uncertainty regarding the function of some members of the family, including bacterial lipocalin Blc from Escherichia coli. Here, we present evidence that lipocalin Blc may be involved in heme binding, trans-periplasmic transport, or heme storage. This conclusion is supported by a cocrystal structure, mass-spectrometric data, absorption titration, and in silico analysis. Binding of heme is observed at low micromolar range with one-to-one ligand-to-protein stoichiometry. However, the absence of classical coordination to the iron atom leaves the possibility that the primary ligand of Blc is another tetrapyrrole.
- Department of Chemistry, Center for Structural Biology, Vanderbilt University, Nashville, TN, USA.
Organizational Affiliation: