Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A
Mascarenhas, R.N., Liu, D., Ballicora, M., Iglesias, A., Asencion, M., Figueroa, C., Esper, M., Aleanzi, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Currently 6VR0 does not have a validation slider image.
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Glucose-1-phosphate adenylyltransferase | 413 | Agrobacterium tumefaciens | Mutation(s): 1 Gene Names: glgC, SY94_4053 EC: 2.7.7.27 | 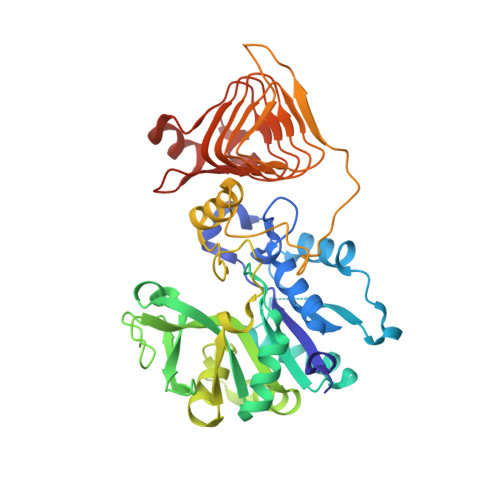 | |
UniProt | |||||
Find proteins for P39669 (Agrobacterium tumefaciens) Explore P39669 Go to UniProtKB: P39669 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P39669 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | AA [auth C] AB [auth F] AC [auth I] BA [auth C] BB [auth F] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | IC [auth I] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 433.318 | α = 90 |
| b = 141.04 | β = 102.3 |
| c = 92.51 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHASER | phasing |
Currently 6VR0 does not have a validation slider image.
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, United States) | United States | MCB 1616851 |