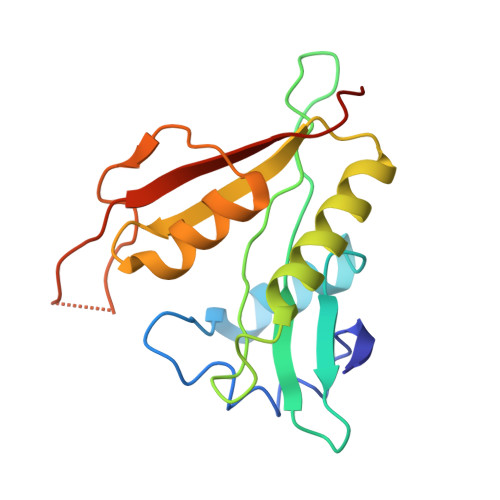
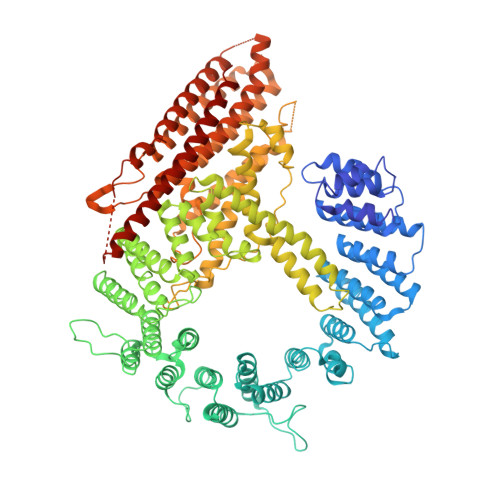
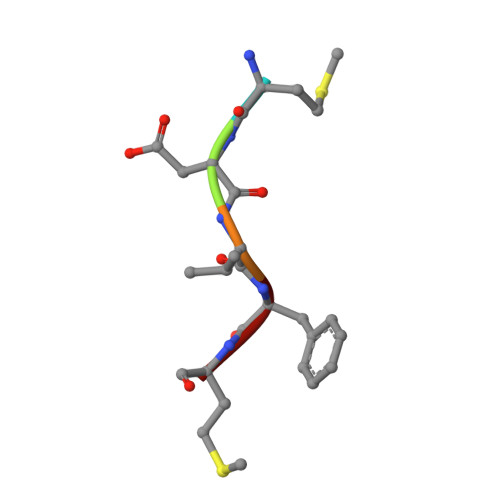
Molecular basis for N-terminal alpha-synuclein acetylation by human NatB.
Deng, S., Pan, B., Gottlieb, L., Petersson, J., Marmorstein, R.(2020) Elife 9
- PubMed: 32885784
- DOI: https://doi.org/10.7554/eLife.57491
- Primary Citation of Related Structures:
6VP9 - PubMed Abstract:
NatB is one of three major N-terminal acetyltransferase (NAT) complexes (NatA-NatC), which co-translationally acetylate the N-termini of eukaryotic proteins. Its substrates account for about 21% of the human proteome, including well known proteins such as actin, tropomyosin, CDK2, and α-synuclein (αSyn). Human NatB (hNatB) mediated N-terminal acetylation of αSyn has been demonstrated to play key roles in the pathogenesis of Parkinson's disease and as a potential therapeutic target for hepatocellular carcinoma. Here we report the cryo-EM structure of hNatB bound to a CoA-αSyn conjugate, together with structure-guided analysis of mutational effects on catalysis. This analysis reveals functionally important differences with human NatA and Candida albicans NatB, resolves key hNatB protein determinants for αSyn N-terminal acetylation, and identifies important residues for substrate-specific recognition and acetylation by NatB enzymes. These studies have implications for developing small molecule NatB probes and for understanding the mode of substrate selection by NAT enzymes.
- Department of Chemistry, University of Pennsylvania, Philadelphia, United States.
Organizational Affiliation: