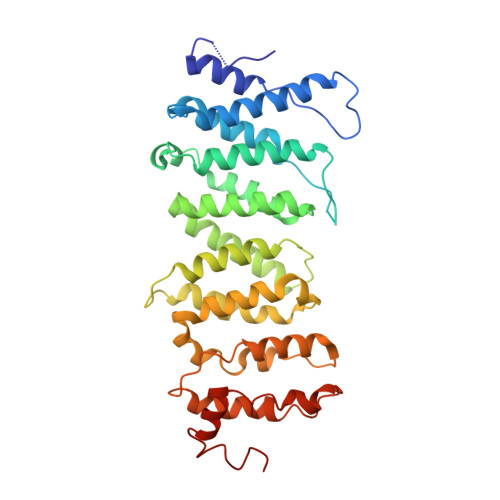
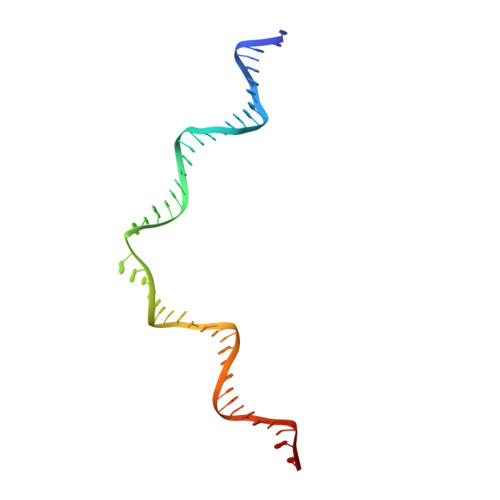
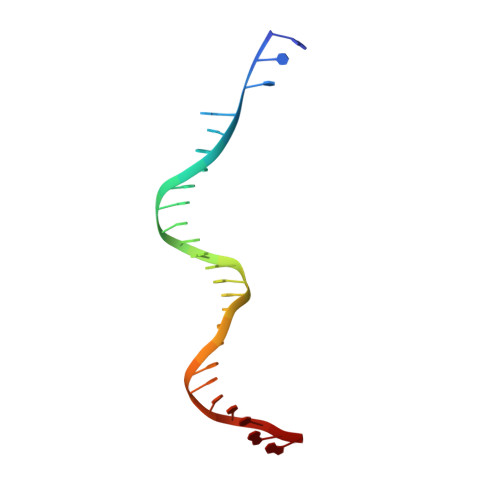
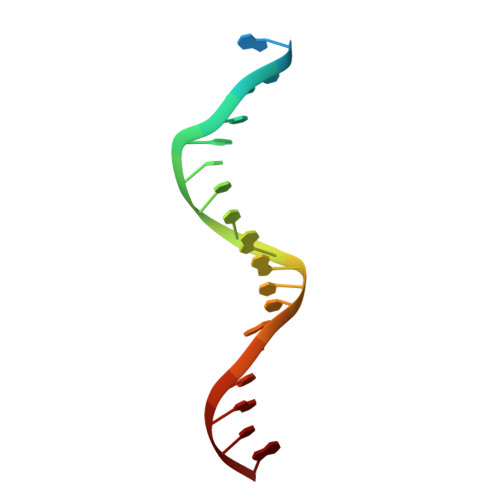
Structural basis of host protein hijacking in human T-cell leukemia virus integration.
Bhatt, V., Shi, K., Salamango, D.J., Moeller, N.H., Pandey, K.K., Bera, S., Bohl, T.E., Kurniawan, F., Orellana, K., Zhang, W., Grandgenett, D.P., Harris, R.S., Sundborger-Lunna, A.C., Aihara, H.(2020) Nat Commun 11: 3121-3121
- PubMed: 32561747
- DOI: https://doi.org/10.1038/s41467-020-16963-6
- Primary Citation of Related Structures:
6VOY - PubMed Abstract:
Integration of the reverse-transcribed viral DNA into host chromosomes is a critical step in the life-cycle of retroviruses, including an oncogenic delta(δ)-retrovirus human T-cell leukemia virus type-1 (HTLV-1). Retroviral integrase forms a higher order nucleoprotein assembly (intasome) to catalyze the integration reaction, in which the roles of host factors remain poorly understood. Here, we use cryo-electron microscopy to visualize the HTLV-1 intasome at 3.7-Å resolution. The structure together with functional analyses reveal that the B56γ (B'γ) subunit of an essential host enzyme, protein phosphatase 2 A (PP2A), is repurposed as an integral component of the intasome to mediate HTLV-1 integration. Our studies reveal a key host-virus interaction underlying the replication of an important human pathogen and highlight divergent integration strategies of retroviruses.
- The Hormel Institute, University of Minnesota, 801 16th Avenue N.E., Austin, MN, 55912, USA.
Organizational Affiliation: