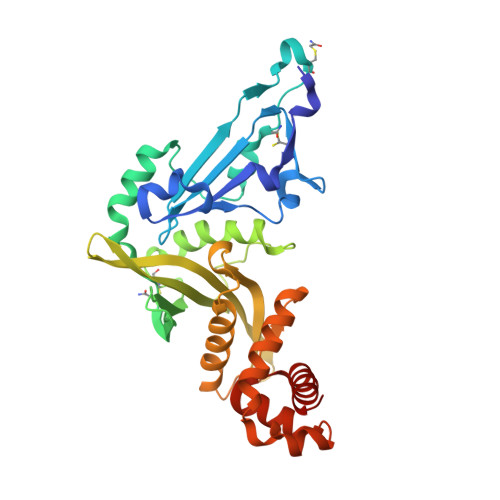
Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4
Halabelian, L., Zeng, H., Dong, A., Loppnau, P., Bountra, C., Edwards, A.M., Arrowsmith, C.H., Structural Genomics Consortium (SGC)To be published.