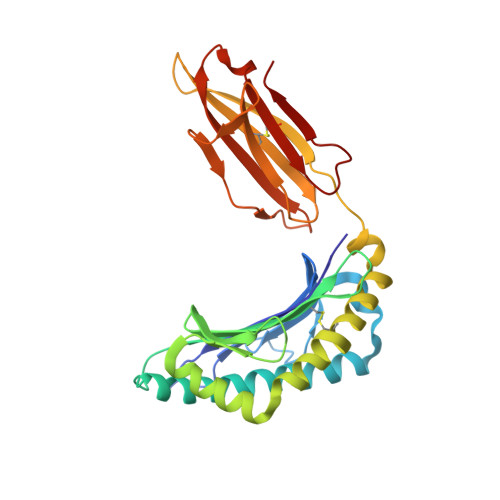
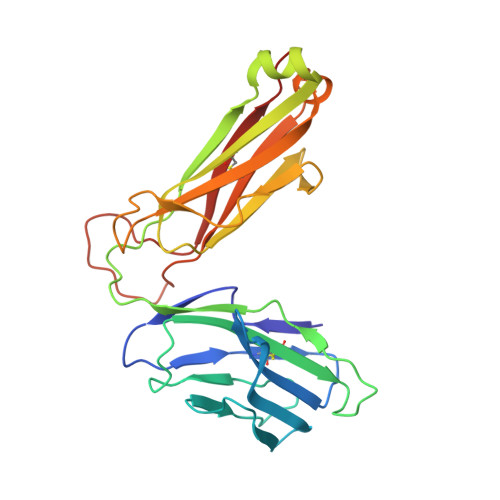
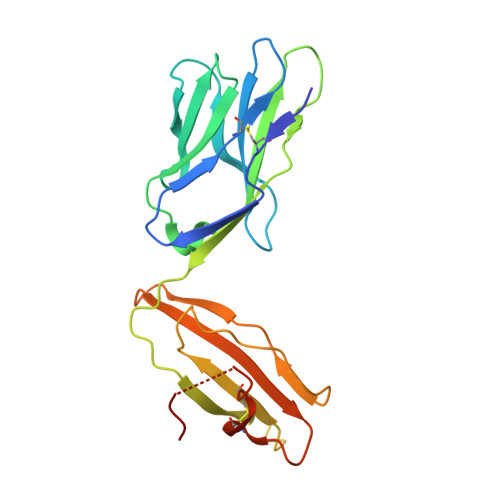
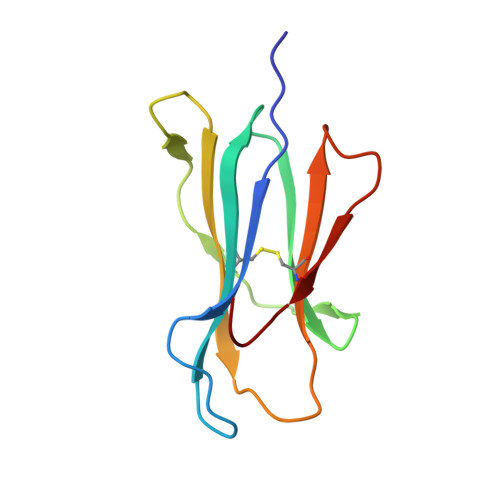
Structurally silent peptide anchor modifications allosterically modulate T cell recognition in a receptor-dependent manner.
Smith, A.R., Alonso, J.A., Ayres, C.M., Singh, N.K., Hellman, L.M., Baker, B.M.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33468649
- DOI: https://doi.org/10.1073/pnas.2018125118
- Primary Citation of Related Structures:
6VM8 - PubMed Abstract:
Presentation of peptides by class I MHC proteins underlies T cell immune responses to pathogens and cancer. The association between peptide binding affinity and immunogenicity has led to the engineering of modified peptides with improved MHC binding, with the hope that these peptides would be useful for eliciting cross-reactive immune responses directed toward their weak binding, unmodified counterparts. Increasing evidence, however, indicates that T cell receptors (TCRs) can perceive such anchor-modified peptides differently than wild-type (WT) peptides, although the scope of discrimination is unclear. We show here that even modifications at primary anchors that have no discernible structural impact can lead to substantially stronger or weaker T cell recognition depending on the TCR. Surprisingly, the effect of peptide anchor modification can be sensed by a TCR at regions distant from the site of modification, indicating a through-protein mechanism in which the anchor residue serves as an allosteric modulator for TCR binding. Our findings emphasize caution in the use and interpretation of results from anchor-modified peptides and have implications for how anchor modifications are accounted for in other circumstances, such as predicting the immunogenicity of tumor neoantigens. Our data also highlight an important need to better understand the highly tunable dynamic nature of class I MHC proteins and the impact this has on various forms of immune recognition.
- Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, IN 46556.
Organizational Affiliation: