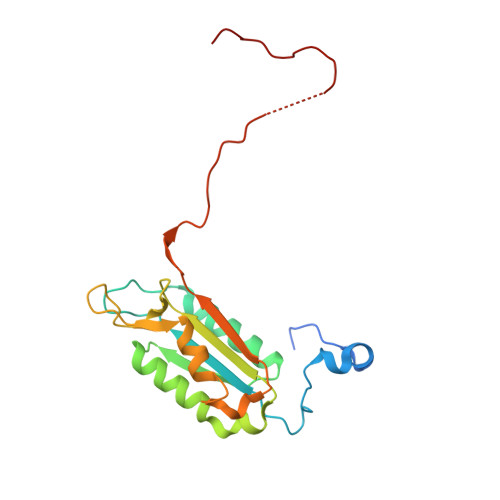
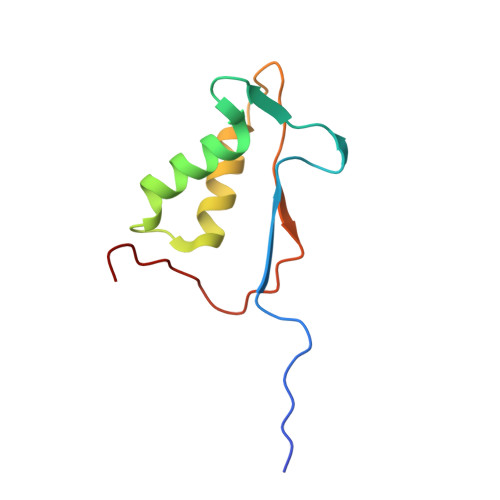
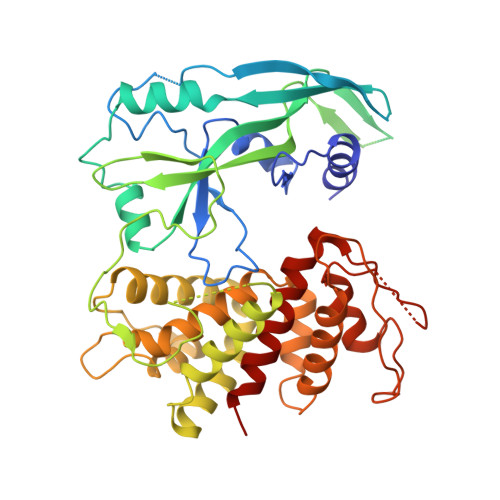
Caspase-1 Engages Full-Length Gasdermin D through Two Distinct Interfaces That Mediate Caspase Recruitment and Substrate Cleavage.
Liu, Z., Wang, C., Yang, J., Chen, Y., Zhou, B., Abbott, D.W., Xiao, T.S.(2020) Immunity 53: 106
- PubMed: 32553275
- DOI: https://doi.org/10.1016/j.immuni.2020.06.007
- Primary Citation of Related Structures:
6VIE - PubMed Abstract:
The recognition and cleavage of gasdermin D (GSDMD) by inflammatory caspases-1, 4, 5, and 11 are essential steps in initiating pyroptosis after inflammasome activation. Previous work has identified cleavage site signatures in substrates such as GSDMD, but it is unclear whether these are the sole determinants for caspase engagement. Here we report the crystal structure of a complex between human caspase-1 and the full-length murine GSDMD. In addition to engagement of the GSDMD N- and C-domain linker by the caspase-1 active site, an anti-parallel β sheet at the caspase-1 L2 and L2' loops bound a hydrophobic pocket within the GSDMD C-terminal domain distal to its N-terminal domain. This "exosite" interface endows an additional function for the GSDMD C-terminal domain as a caspase-recruitment module besides its role in autoinhibition. Our study thus reveals dual-interface engagement of GSDMD by caspase-1, which may be applicable to other physiological substrates of caspases.
- Department of Pathology, Case Western Reserve University, Cleveland, OH 44106, USA.
Organizational Affiliation: