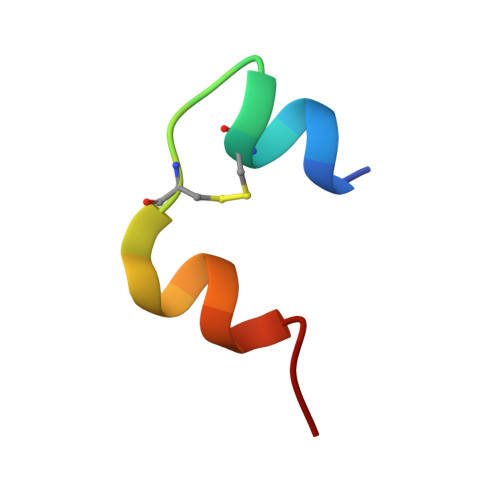
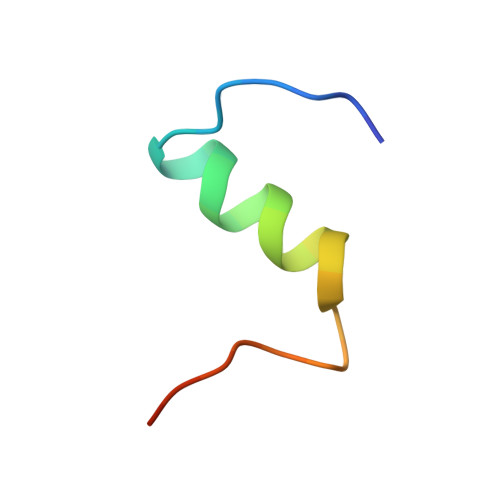
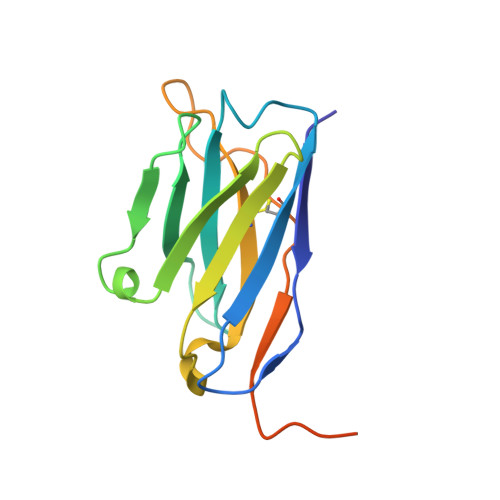
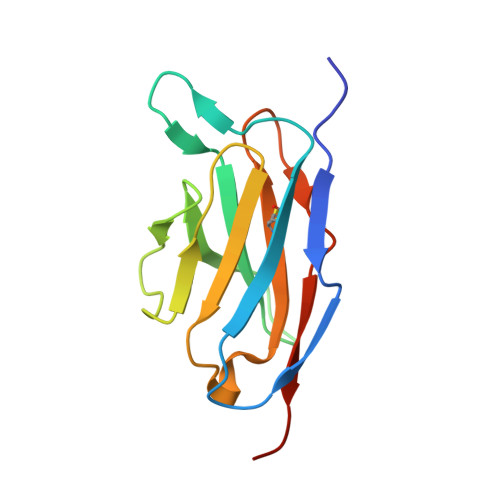
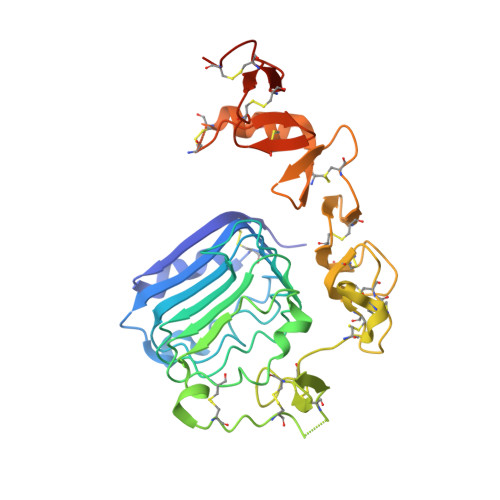
A structurally minimized yet fully active insulin based on cone-snail venom insulin principles.
Xiong, X., Menting, J.G., Disotuar, M.M., Smith, N.A., Delaine, C.A., Ghabash, G., Agrawal, R., Wang, X., He, X., Fisher, S.J., MacRaild, C.A., Norton, R.S., Gajewiak, J., Forbes, B.E., Smith, B.J., Safavi-Hemami, H., Olivera, B., Lawrence, M.C., Chou, D.H.(2020) Nat Struct Mol Biol 27: 615-624
- PubMed: 32483339
- DOI: https://doi.org/10.1038/s41594-020-0430-8
- Primary Citation of Related Structures:
6VEP, 6VEQ, 6VET - PubMed Abstract:
Human insulin and its current therapeutic analogs all show propensity, albeit varyingly, to self-associate into dimers and hexamers, which delays their onset of action and makes blood glucose management difficult for people with diabetes. Recently, we described a monomeric, insulin-like peptide in cone-snail venom with moderate human insulin-like bioactivity. Here, with insights from structural biology studies, we report the development of mini-Ins-a human des-octapeptide insulin analog-as a structurally minimal, full-potency insulin. Mini-Ins is monomeric and, despite the lack of the canonical B-chain C-terminal octapeptide, has similar receptor binding affinity to human insulin. Four mutations compensate for the lack of contacts normally made by the octapeptide. Mini-Ins also has similar in vitro insulin signaling and in vivo bioactivities to human insulin. The full bioactivity of mini-Ins demonstrates the dispensability of the PheB24-PheB25-TyrB26 aromatic triplet and opens a new direction for therapeutic insulin development.
- Department of Biochemistry, University of Utah, Salt Lake City, UT, USA.
Organizational Affiliation: