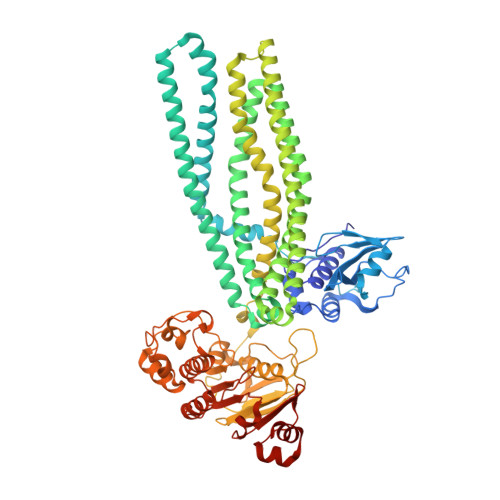
Structural basis of substrate recognition by a polypeptide processing and secretion transporter.
Kieuvongngam, V., Olinares, P.D.B., Palillo, A., Oldham, M.L., Chait, B.T., Chen, J.(2020) Elife 9
- PubMed: 31934861
- DOI: https://doi.org/10.7554/eLife.51492
- Primary Citation of Related Structures:
6V9Z - PubMed Abstract:
The peptidase-containing ATP-binding cassette transporters (PCATs) are unique members of the ABC transporter family that proteolytically process and export peptides and proteins. Each PCAT contains two peptidase domains that cleave off the secretion signal, two transmembrane domains forming a translocation pathway, and two nucleotide-binding domains that hydrolyze ATP. Previously the crystal structures of a PCAT from Clostridium thermocellum (PCAT1) were determined in the absence and presence of ATP, revealing how ATP binding regulates the protease activity and access to the translocation pathway. However, how the substrate CtA, a 90-residue polypeptide, is recognized by PCAT1 remained elusive. To address this question, we determined the structure of the PCAT1-CtA complex by electron cryo-microscopy (cryo-EM) to 3.4 Å resolution. The structure shows that two CtAs are bound via their N-terminal leader peptides, but only one is positioned for cleavage and translocation. Based on these results, we propose a model of how substrate cleavage, ATP hydrolysis, and substrate translocation are coordinated in a transport cycle.
- Laboratory of Membrane Biophysics and Biology, The Rockefeller University, New York, United States.
Organizational Affiliation: