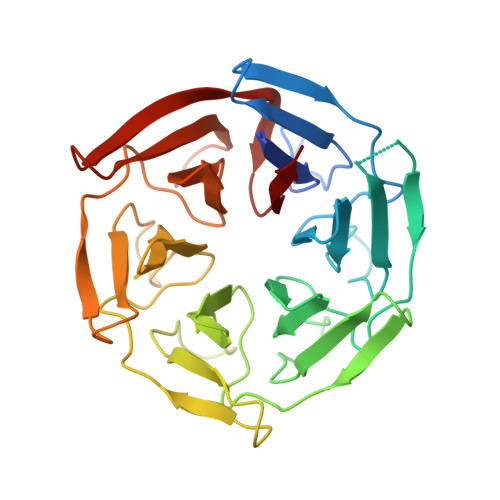
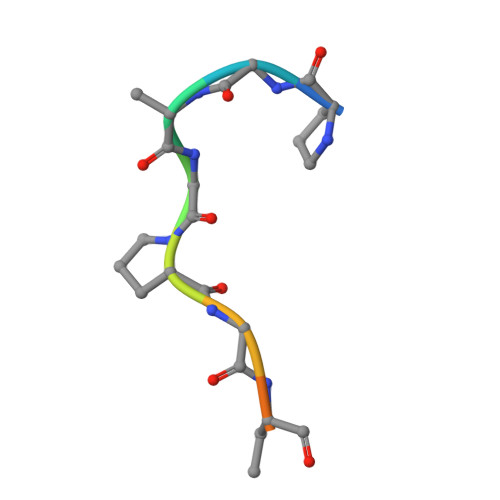
Structural Elucidation of Peptide Binding to KLHL-12, a Substrate Specific Adapter Protein in a Cul3-Ring E3 Ligase Complex.
Zhao, B., Payne, W.G., Sai, J., Lu, Z., Olejniczak, E.T., Fesik, S.W.(2020) Biochemistry 59: 964-969
- PubMed: 32032490
- DOI: https://doi.org/10.1021/acs.biochem.9b01073
- Primary Citation of Related Structures:
6V7O - PubMed Abstract:
KLHL-12 is a substrate specific adapter protein for a Cul3-Ring ligase complex. It is a member of the Kelch β-propeller domain subclass of Cullin-Ring substrate recognition domains. This E3 ubiquitin ligase complex has many activities, including acting as a negative regulator of the Wnt signaling pathway by mediating ubiquitination and subsequent proteolysis of Dvl3/Dsh3. KLHL-12 is also known to mediate the polyubiquitination of the dopamine D4 receptor (D4.2), the ubiquitination of KHSRP, a protein that is involved in IRES translation, and also the ubiquitination of Sec31, which is involved in endoplasmic reticulum-Golgi transport by regulating the size of COPII coats. Earlier studies broadly defined the substrate binding regions for D4.2 and Dvl3/Dsh3 to KLHL-12. We tested several peptides from these regions and succeeded in identifying a short peptide that bound to KLHL-12 with low micromolar affinity. To better understand the sequence specificity of this peptide, we used alanine substitutions to map the important residues and obtained an X-ray structure of this peptide bound to KLHL-12. This structure and our peptide affinity measurements suggest a sequence motif for peptides that bind to the top face of KLHL-12. Understanding this binding site on KLHL-12 may contribute to efforts to find small molecule ligands that can either directly inhibit the degradation of substrate proteins or be used in targeted protein degradation strategies using PROTACs.
- Department of Biochemistry, Vanderbilt University School of Medicine, 2215 Garland Avenue, 607 Light Hall, Nashville, Tennessee 37232-0146, United States.
Organizational Affiliation: