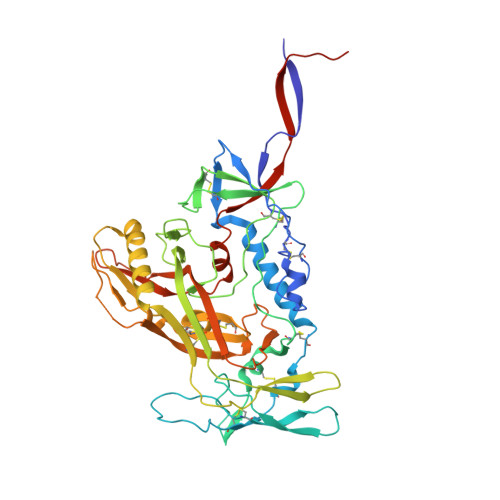
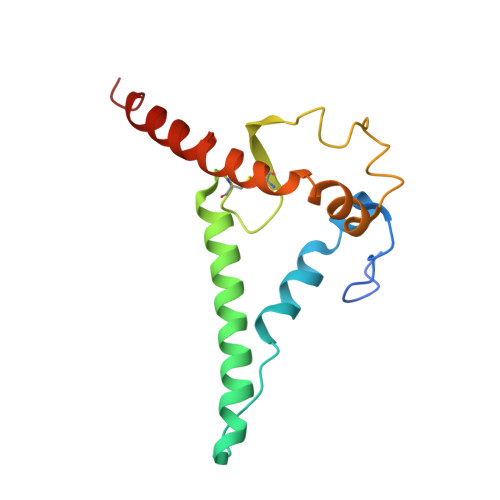
A VH1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite
Kumar, S., Ju, B., Shapero, B., Lin, X., Ren, L., Zhang, L., Li, D., Zhou, Z., Feng, Y., Sou, C., Mann, C.J., Hao, Y., Sarkar, A., Hou, J., Nunnally, C., Hong, K., Wang, S., Ge, X., Su, B., Landais, E., Sok, D., Zwick, M.B., He, L., Zhu, J., Wilson, I.A., Shao, Y.(2020) Sci Adv 6: eabb1328