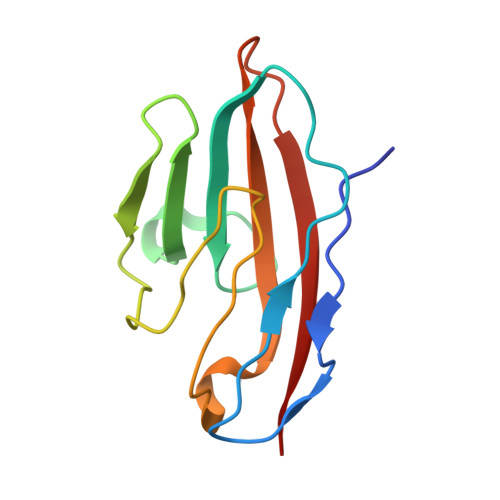
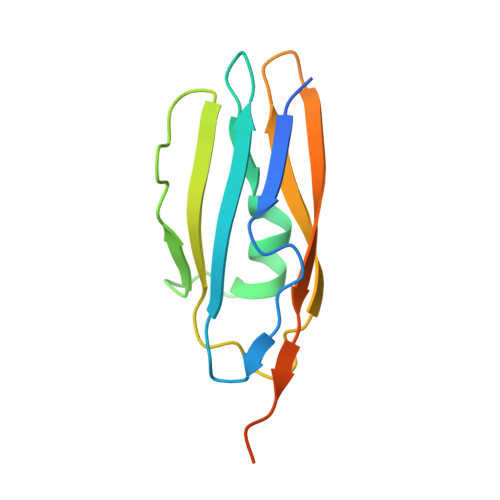
Bacterial protein domains with a novel Ig-like fold target human CEACAM receptors.
van Sorge, N.M., Bonsor, D.A., Deng, L., Lindahl, E., Schmitt, V., Lyndin, M., Schmidt, A., Nilsson, O.R., Brizuela, J., Boero, E., Sundberg, E.J., van Strijp, J.A.G., Doran, K.S., Singer, B.B., Lindahl, G., McCarthy, A.J.(2021) EMBO J 40: e106103-e106103
- PubMed: 33522633
- DOI: https://doi.org/10.15252/embj.2020106103
- Primary Citation of Related Structures:
6V3P - PubMed Abstract:
Streptococcus agalactiae, also known as group B Streptococcus (GBS), is the major cause of neonatal sepsis in humans. A critical step to infection is adhesion of bacteria to epithelial surfaces. GBS adhesins have been identified to bind extracellular matrix components and cellular receptors. However, several putative adhesins have no host binding partner characterised. We report here that surface-expressed β protein of GBS binds to human CEACAM1 and CEACAM5 receptors. A crystal structure of the complex showed that an IgSF domain in β represents a novel Ig-fold subtype called IgI3, in which unique features allow binding to CEACAM1. Bioinformatic assessment revealed that this newly identified IgI3 fold is not exclusively present in GBS but is predicted to be present in adhesins from other clinically important human pathogens. In agreement with this prediction, we found that CEACAM1 binds to an IgI3 domain found in an adhesin from a different streptococcal species. Overall, our results indicate that the IgI3 fold could provide a broadly applied mechanism for bacteria to target CEACAMs.
- Department of Medical Microbiology, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Organizational Affiliation: