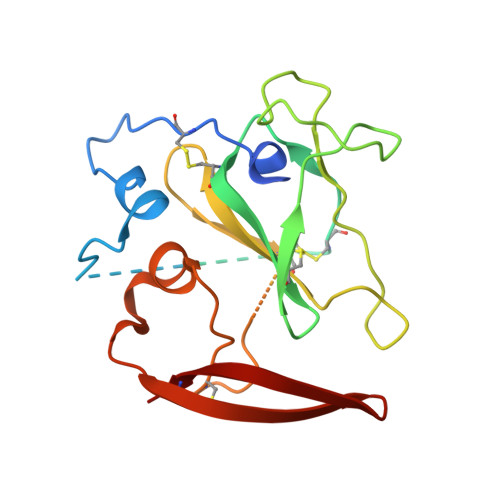
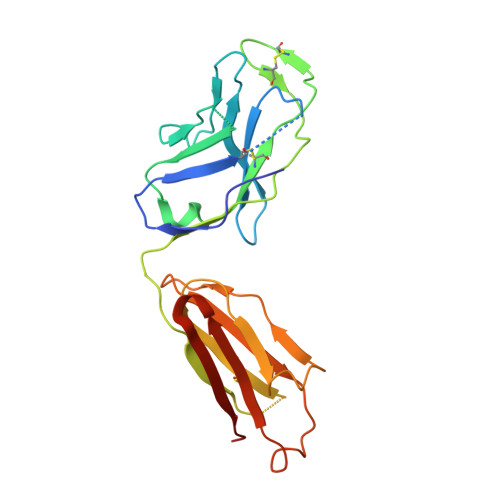
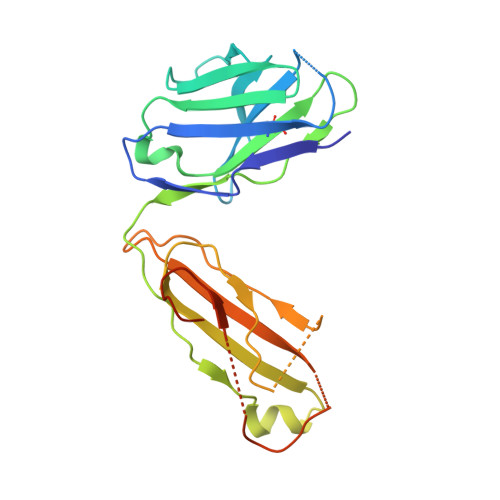
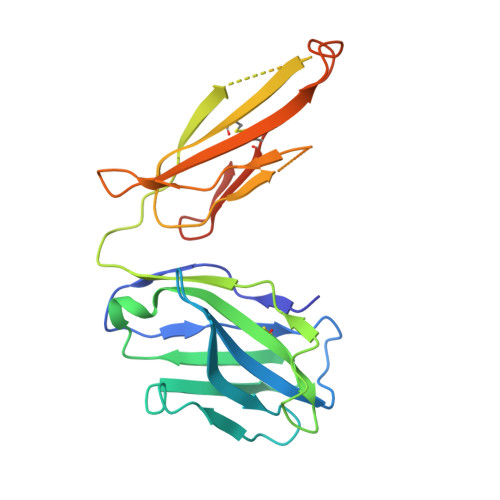
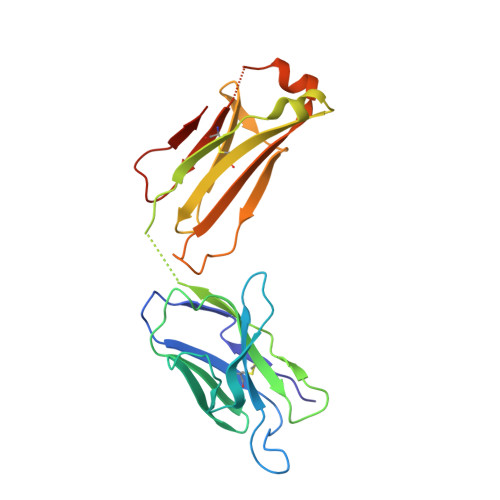
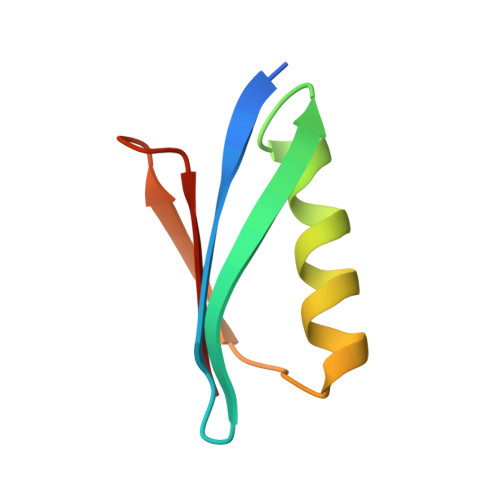
Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
He, L., Tzarum, N., Lin, X., Shapero, B., Sou, C., Mann, C.J., Stano, A., Zhang, L., Nagy, K., Giang, E., Law, M., Wilson, I.A., Zhu, J.(2020) Sci Adv 6: eaaz6225-eaaz6225
- PubMed: 32494617
- DOI: https://doi.org/10.1126/sciadv.aaz6225
- Primary Citation of Related Structures:
6UYD, 6UYF, 6UYG, 6UYM - PubMed Abstract:
Hepatitis C virus (HCV) envelope glycoproteins E1 and E2 are responsible for cell entry, with E2 being the major target of neutralizing antibodies (NAbs). Here, we present a comprehensive strategy for B cell-based HCV vaccine development through E2 optimization and nanoparticle display. We redesigned variable region 2 in a truncated form (tVR2) on E2 cores derived from genotypes 1a and 6a, resulting in improved stability and antigenicity. Crystal structures of three optimized E2 cores with human cross-genotype NAbs (AR3s) revealed how the modified tVR2 stabilizes E2 without altering key neutralizing epitopes. We then displayed these E2 cores on 24- and 60-meric nanoparticles and achieved substantial yield and purity, as well as enhanced antigenicity. In mice, these nanoparticles elicited more effective NAb responses than soluble E2 cores. Next-generation sequencing (NGS) defined distinct B cell patterns associated with nanoparticle-induced antibody responses, which target the conserved neutralizing epitopes on E2 and cross-neutralize HCV genotypes.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: