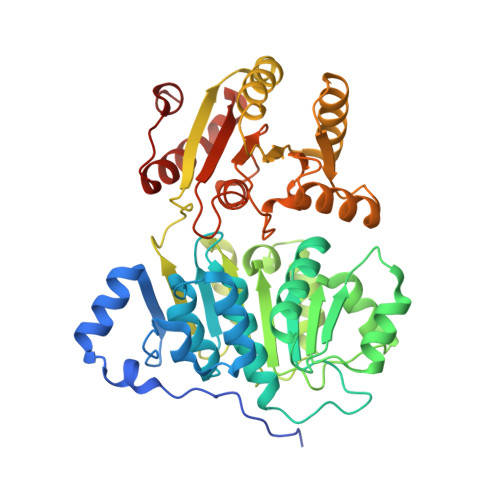
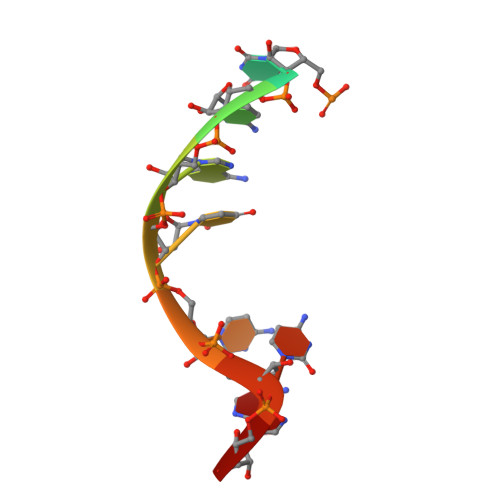
RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Ngo, T.D., Partin, A.C., Nam, Y.(2019) Cell Rep 29: 4024-4035.e5
- PubMed: 31851931
- DOI: https://doi.org/10.1016/j.celrep.2019.11.059
- Primary Citation of Related Structures:
6UV0, 6UV1, 6UV2, 6UV3, 6UV4 - PubMed Abstract:
DDX17, a DEAD-box ATPase, is a multifunctional helicase important for various RNA functions, including microRNA maturation. Key questions for DDX17 include how it recognizes target RNAs and influences their structures, as well as how its ATPase activity may be regulated. Through crystal structures and biochemical assays, we show the ability of the core catalytic domains of DDX17 to recognize specific sequences in target RNAs. The RNA sequence preference of the catalytic core is critical for DDX17 to directly bind and remodel a specific region of primary microRNAs 3' to the mature sequence, and consequently enhance processing by Drosha. Furthermore, we identify an intramolecular interaction between the N-terminal tail and the DEAD domain of DDX17 to have an autoregulatory role in controlling the ATPase activity. Thus, we provide the molecular basis for how cognate RNA recognition and functional outcomes are linked for DDX17.
- Cecil H. and Ida Green Center for Reproductive Biology Sciences and Division of Basic Reproductive Biology Research, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA; Department of Biophysics, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA; Department of Obstetrics and Gynecology, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: