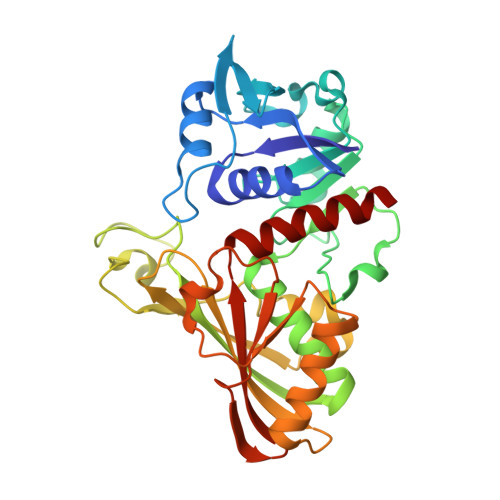
A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Rodriguez-Hernandez, A., Romo-Arevalo, E., Rodriguez-Romero, A.(2019) Crystals (Basel) 9: 622