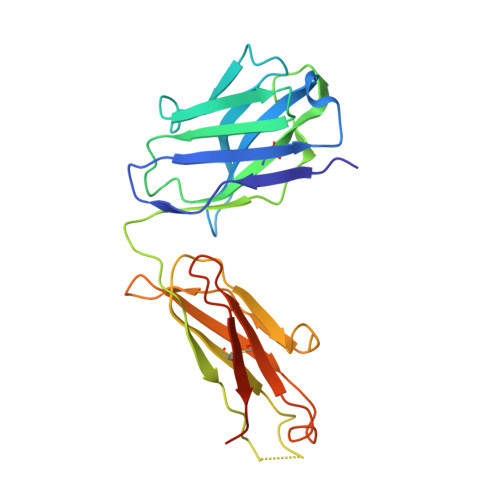
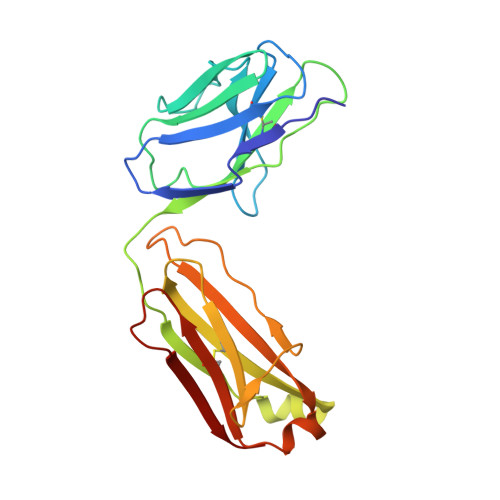
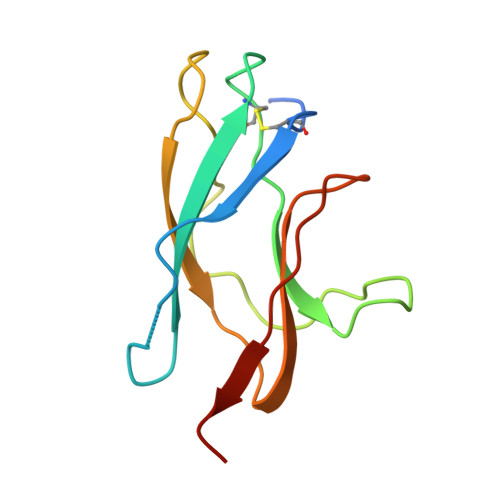
Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Esswein, S.R., Gristick, H.B., Jurado, A., Peace, A., Keeffe, J.R., Lee, Y.E., Voll, A.V., Saeed, M., Nussenzweig, M.C., Rice, C.M., Robbiani, D.F., MacDonald, M.R., Bjorkman, P.J.(2020) Proc Natl Acad Sci U S A 117: 9865-9875
- PubMed: 32321830
- DOI: https://doi.org/10.1073/pnas.1919269117
- Primary Citation of Related Structures:
6UTA, 6UTE - PubMed Abstract:
Recent epidemics demonstrate the global threat of Zika virus (ZIKV), a flavivirus transmitted by mosquitoes. Although infection is usually asymptomatic or mild, newborns of infected mothers can display severe symptoms, including neurodevelopmental abnormalities and microcephaly. Given the large-scale spread, symptom severity, and lack of treatment or prophylaxis, a safe and effective ZIKV vaccine is urgently needed. However, vaccine design is complicated by concern that elicited antibodies (Abs) may cross-react with other flaviviruses that share a similar envelope protein, such as dengue virus, West Nile virus, and yellow fever virus. This cross-reactivity may worsen symptoms of a subsequent infection through Ab-dependent enhancement. To better understand the neutralizing Ab response and risk of Ab-dependent enhancement, further information on germline Ab binding to ZIKV and the maturation process that gives rise to potently neutralizing Abs is needed. Here we use binding and structural studies to compare mature and inferred-germline Ab binding to envelope protein domain III of ZIKV and other flaviviruses. We show that affinity maturation of the light-chain variable domain is important for strong binding of the recurrent VH3-23/VK1-5 neutralizing Abs to ZIKV envelope protein domain III, and identify interacting residues that contribute to weak, cross-reactive binding to West Nile virus. These findings provide insight into the affinity maturation process and potential cross-reactivity of VH3-23/VK1-5 neutralizing Abs, informing precautions for protein-based vaccines designed to elicit germline versions of neutralizing Abs.
- Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA 91125.
Organizational Affiliation: