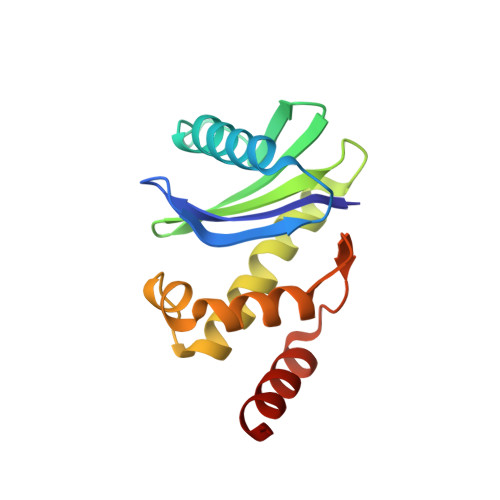
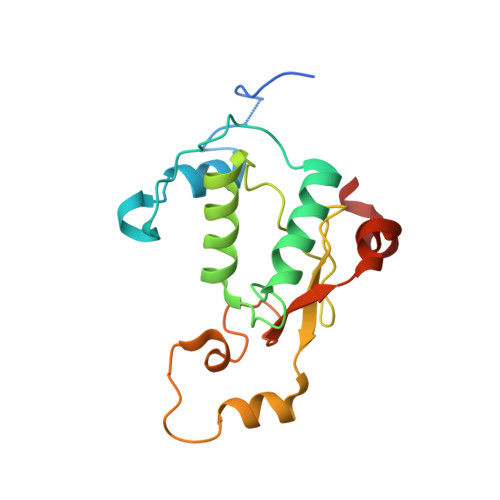
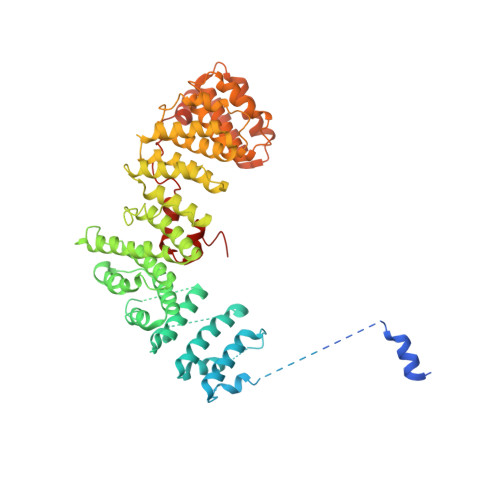
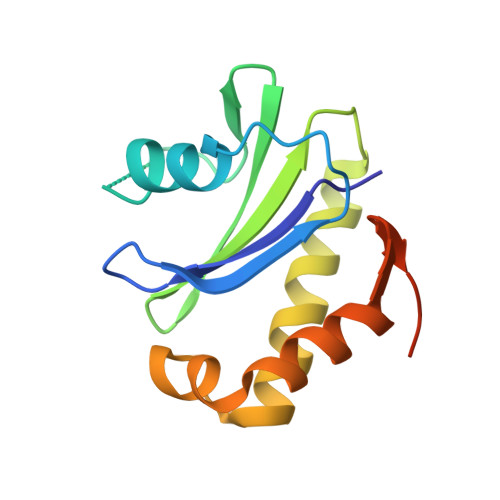
Structural basis of CD4 downregulation by HIV-1 Nef.
Kwon, Y., Kaake, R.M., Echeverria, I., Suarez, M., Karimian Shamsabadi, M., Stoneham, C., Ramirez, P.W., Kress, J., Singh, R., Sali, A., Krogan, N., Guatelli, J., Jia, X.(2020) Nat Struct Mol Biol 27: 822-828
- PubMed: 32719457
- DOI: https://doi.org/10.1038/s41594-020-0463-z
- Primary Citation of Related Structures:
6URI, 9A0E - PubMed Abstract:
The HIV-1 Nef protein suppresses multiple immune surveillance mechanisms to promote viral pathogenesis and is an attractive target for the development of novel therapeutics. A key function of Nef is to remove the CD4 receptor from the cell surface by hijacking clathrin- and adaptor protein complex 2 (AP2)-dependent endocytosis. However, exactly how Nef does this has been elusive. Here, we describe the underlying mechanism as revealed by a 3.0-Å crystal structure of a fusion protein comprising Nef and the cytoplasmic domain of CD4 bound to the tetrameric AP2 complex. An intricate combination of conformational changes occurs in both Nef and AP2 to enable CD4 binding and downregulation. A pocket on Nef previously identified as crucial for recruiting class I MHC is also responsible for recruiting CD4, revealing a potential approach to inhibit two of Nef's activities and sensitize the virus to immune clearance.
- Department of Chemistry and Biochemistry, University of Massachusetts Dartmouth, Dartmouth, MA, USA.
Organizational Affiliation: