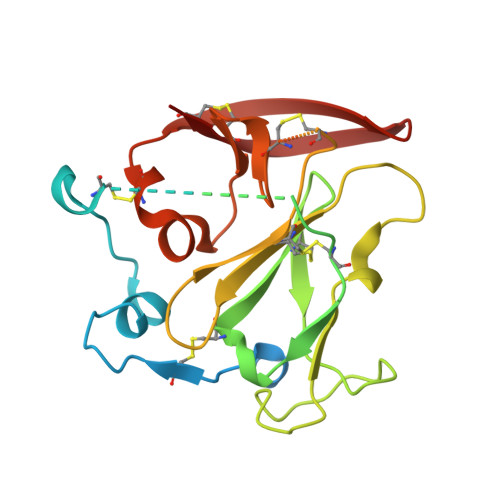
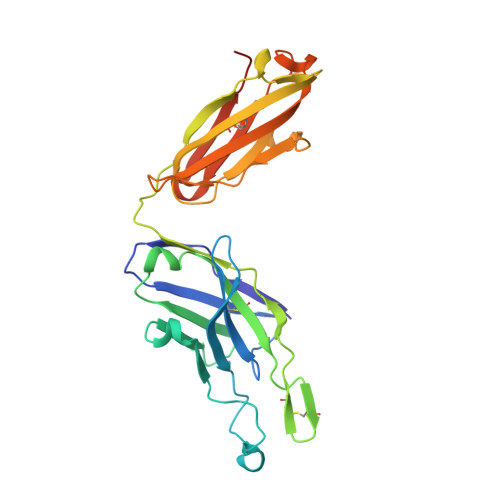
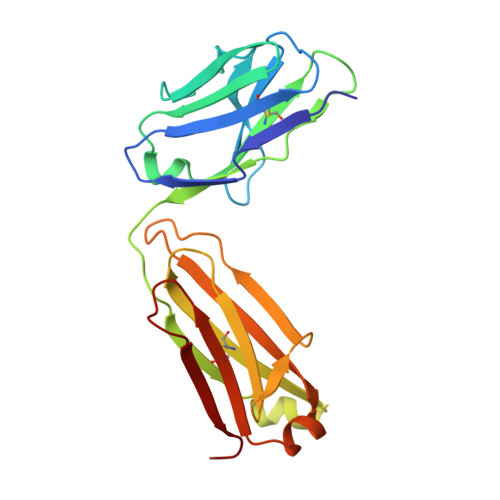
An ultralong CDRH2 in HCV neutralizing antibody demonstrates structural plasticity of antibodies against E2 glycoprotein.
Flyak, A.I., Ruiz, S.E., Salas, J., Rho, S., Bailey, J.R., Bjorkman, P.J.(2020) Elife 9
- PubMed: 32125272
- DOI: https://doi.org/10.7554/eLife.53169
- Primary Citation of Related Structures:
6URH - PubMed Abstract:
A vaccine protective against diverse HCV variants is needed to control the HCV epidemic. Structures of E2 complexes with front layer-specific broadly neutralizing antibodies (bNAbs) isolated from HCV-infected individuals, revealed a disulfide bond-containing CDRH3 that adopts straight (individuals who clear infection) or bent (individuals with chronic infection) conformation. To investigate whether a straight versus bent disulfide bond-containing CDRH3 is specific to particular HCV-infected individuals, we solved a crystal structure of the HCV E2 ectodomain in complex with AR3X, a bNAb with an unusually long CDRH2 that was isolated from the chronically-infected individual from whom the bent CDRH3 bNAbs were derived. The structure revealed that AR3X utilizes both its ultralong CDRH2 and a disulfide motif-containing straight CDRH3 to recognize the E2 front layer. These results demonstrate that both the straight and bent CDRH3 classes of HCV bNAb can be elicited in a single individual, revealing a structural plasticity of VH1-69 -derived bNAbs.
- Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, United States.
Organizational Affiliation: